MarkUs
Contents
Overview
MarkUs is a web server to assist the assessment of the biochemical function for a given protein structure. MarkUs identifies related protein structures and sequences, detects protein cavities, and calculates the surface electrostatic potentials and amino acid conservation profile. The results can be browsed by an interactive web interface that allows one to integrate Gene Ontology terms, UniProt features, and the Enzyme Classification. The MarkUs website is at https://honiglab.c2b2.columbia.edu/MarkUs/cgi-bin/submit.pl.
geWorkbench previously supported both a direct link to the MarkUs server via a web service, and also a connection through a grid service. However, the grid service is no longer active. No password is required for either the web or grid service interface to MarkUs.
Availability of results
MarkUs results are returned on demand from the server into the browser window in geWorkbench. The results will be available within geWorkbench for as long as they are preserved on the server, which should be at least 30 days. It is not possible to preserve the entire MarkUs run result within geWorkbench. Hence, after a MarkUs run, the relevant results should be determined and saved to an appropriate format (screenshot, etc).
To maintain access to a particular MarkUs result, you should either save the Workspace before exiting geWorkbench, or make note of the MarkUs job ID. It can be used to retrieve the results at a later time.
Java Requirements for Viewing MarkUs Results
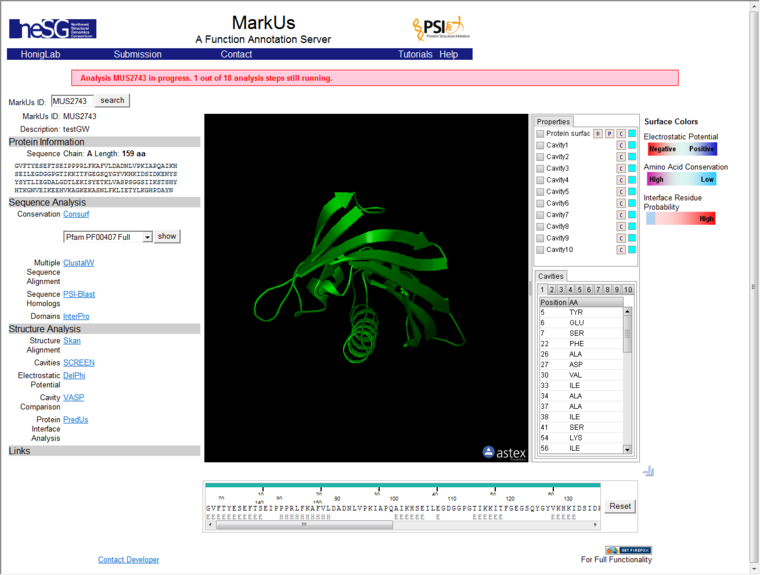
MarkUs results will be displayed either directly in geWorkbench, using a built-in browser, or in the case where a 64-bit Java Virtual Machine is in use (now the default on all platforms) the user can click a link to display the results page in an external web browser window.
The MarkUs results page includes the Astex molecular structure viewer, which is a Java applet. Allowing this applet to run in a browser requires several steps. These are described below.
Java JRE Version
The molecular structure display works in any modern Java version, including Java 7 and Java 8 JREs. You should install the latest version available.
A JRE must be installed which matches the architecture (32 or 64-bit) of the browser being used.
Particular versions of the Java JRE can be downloaded from http://java.oracle.com. 32-bit versions of the JRE include "x86" in their descriptions and "i586" in their file names.
Windows---
As of early 2015, on Windows, if you wish to use the Java JRE in Firefox or the default version of Chrome (both of which are 32-bit browsers), you must install a 32-bit version of the JRE. This may also apply to other browsers. You can have both 32 and 64 bit versions of Java installed on the same machine, if you have a 64-bit version of the operation system.
Mac OS X--- On Mac OS X, the Astex applet should run in Firefox, Chrome or Safari. See Java 7 System Requirements for further information.
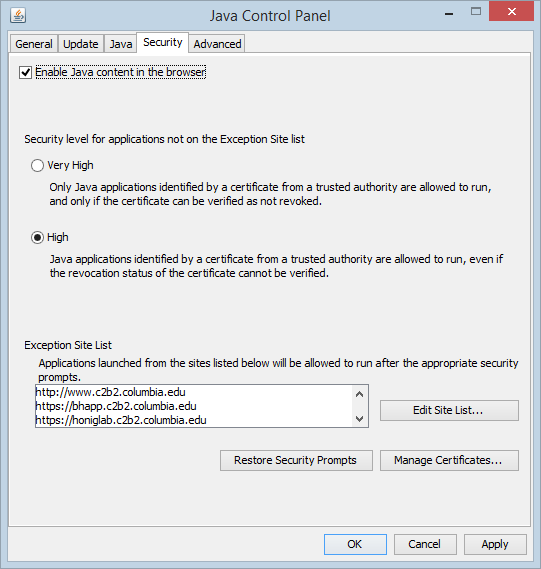
Java Security
To use the Astex molecular structure viewer in the browser, you will need to allow Java to run in the browser, and enable the MarkUs URLs.
On Windows, this can be done by going to Control Panel -> Java -> Security. On the Mac, go to System Preferences -> Java -> Security.
Once in the Java Security tab, two steps are needed:
- Check the box "Enable Java content in the browser"
- Add an exception to allow Java to run for a particular URL.
- Click "Edit Site List" and then "Add".
- Type in these URLs:
- Then click the "OK" button.
Browser Security
Your web browser will also ask for permission to run the viewer applet (Astex). In general, you can choose to either allow it to run only this one time, or to always allow it to run. This choice is up to the user.
MarkUs Server Documentation
Direct Submission of a job to the MarkUs server
https://honiglab.c2b2.columbia.edu/MarkUs/cgi-bin/submit.pl
This page contains a concise description of the goals of the MarkUs server and a link to the direct web job submission page.
MarkUs Server NESG tutorial
https://bhapp.c2b2.columbia.edu/nesg/documentation/tutorial.html
Step by step illustration of submitting a job and interpreting the results.
Detailed Overview of MarkUs Analysis
https://bhapp.c2b2.columbia.edu/nesg/documentation/index.html
Describes each aspect of the Markus analysis results and their presentation.
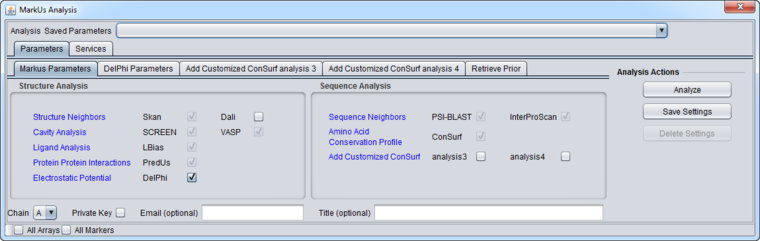
Parameters
Main
- Private Key - if checked, the system will generate and displayed the key in the name of the result node. The private key will be needed to retrieve the job results.
- Email (optional) - enter an Email address to request notification when the job has finished.
- Title (optional) - provide a title for your job.
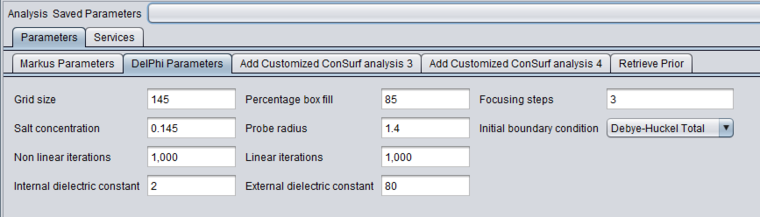
DelPhi
ConSurf
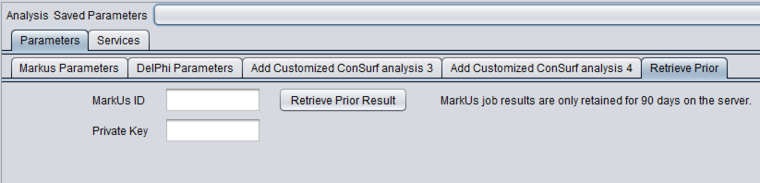
Retrieve Prior Jobs
The results of previously run MarkUs jobs may be retrieved from the MarkUs server at a later time. However, note that results may be deleted after 90 days.
To retrieve prior results, you must have, at the time the job was submitted, made a note of the job id and, if one was requested, the private key (see section below on the Private Key).
- MarkUs ID - When a MarkUs job is submitted, a job ID is automatically generated by the MarkUs server. This ID is displayed in the results page, and is also used as the results node name in the Workspace.
- Private Key - If the "Private Key" box was checked on the Parameters tab when the job was submitted, the system also generated and displayed this key in the name of the result node. In this case, you must also supply this private key to retrieve the job results.
Results
Private Key
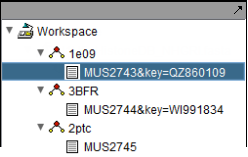
The figure below depicts three separate runs of MarkUs. In the first two, the "Private Key" checkbox was checked in the Parameters tab. The result node name in this case, e.g. "MUS2743&key=QZ860109" contains both the job name, e.g. MUS2743, as well as the private key following the label "key=", in this case "QZ860109".
For jobs which requested a private key, you will need to supply that key in the "Retrieve Prior" tab if you wish to view the results in another invocation of geWorkbench.
Result Viewer
MarkUs results will be displayed either directly in geWorkbench, using a built-in browser, or in the case where a 64-bit Java Virtual Machine is in use (now the default on all platforms) the user can click a link to display the results page in an external web browser window.
The run may take a considerable amount of time to finish, and partial results will be displayed. The window does not auto-refresh. To see the most recent results, right-click on the result display window and select "refresh". A banner will indicate how many steps have yet to be finished for the current run.
Before the first results are available, the results page may display a field for entering a Private Key. If a private key was requested, it has already been generated and is available in the Workspace data node name. This field will disappear once the the first results area available. You do not need to enter anything into this field in the results viewer.
- Technical Note - In the web version of MarkUs, if a private key was requested, it is immediately returned in a preliminary web page. geWorkbench does not display this page.
Running MarkUs
The starting point for a MarkUs analysis is a PDB protein structure file.
1. If the MarkUs component has not been loaded in the Component Configuration Manager (CCM), first do so.
2. Load a PDB file for the protein whose structure you would like to analyze.
3. Set the desired parameters. Note that some analyses have check-boxes next to them which are grayed-out. These analyses are always run as part of a MarkUs job and cannot be left out.
4. If the "private key" checkbox is checked, MarkUs will generate a private key which will be displayed in the result node in the Workspace.
5. Inspect and adjust if desired the parameters to DelPhi. DelPhi calculates the electrostatic field around the molecule.
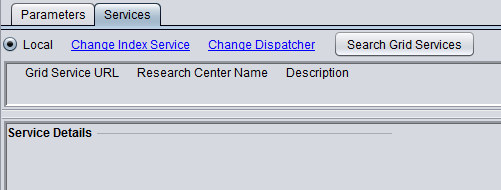
6. MarkUs is available in geWorkbench only as a local service. The grid service has been discontinued.
The figure below shows the local service as selected.
7. Return to the Parameters tab.
8. Push the "Analyze" button.
9. A result node will be created in the Workspace.
10. If the "Private Key" checkbox was checked, copy down the private key from the new result node in the Workspace.
References
Fischer M, Zhang QC, Dey F, Chen BY, Honig B, Petrey D.(2011) MarkUs: a server to navigate sequence-structure-function space. Nucleic Acids Res. Jul;39(Web Server issue):W357-61. Epub 2011 Jun 14. link