Pudge
Overview
The Pudge component serves as a front end to the web-based Pudge protein structure modeling service developed by the Honig lab at Columbia University.
Computational protein structure prediction using sequence homology is a process that exploits the observation that proteins with similar sequences usually have similar structures as well. While this idea is simple in theory, in practice, the process of using sequence homology to predict protein structure is intricate and complex. The process is generally divided into five stages, involving the use of a combination of techniques from a variety of scientific disciplines. See Petrey and Honig, 2005 for a description of the overall protein structure prediction process.
External Tutorial
The Honig lab has provided their own tutorial for Pudge.
In addition, Pudge can be run directly on their web server.
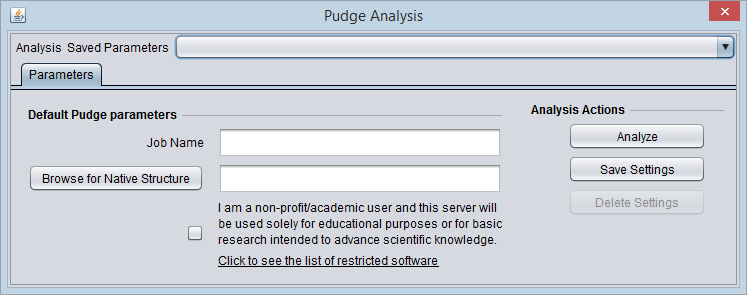
Parameters and running Pudge
If Pudge has been loaded via the CCM, it will appear in the Analysis area when a FASTA sequence file is loaded and selected in the Workspace.
To run Pudge,
1. Job Name - add a job name in the Job Name text field.
2. Browse for Native Structure (optional) - this is not currently working. The intended use was to add a PDB protein structure file to serve as a starting structural template.
Academic message: certain methods used in Pudge are restricted to academic users. Substitutes are offered. If you are an academic user, please check the box.
Hovering the cursor over the "restricted software" link will display a more detailed description:
3. Click Analyze - In the PudgeBrowser tab, the Pudge Analysis interface will be displayed either directly in geWorkbench, using a built-in browser, or in the case where a 64-bit Java Virtual Machine is in use (now the default on all platforms) the user can click a link to display the analysis page in an external web browser window.
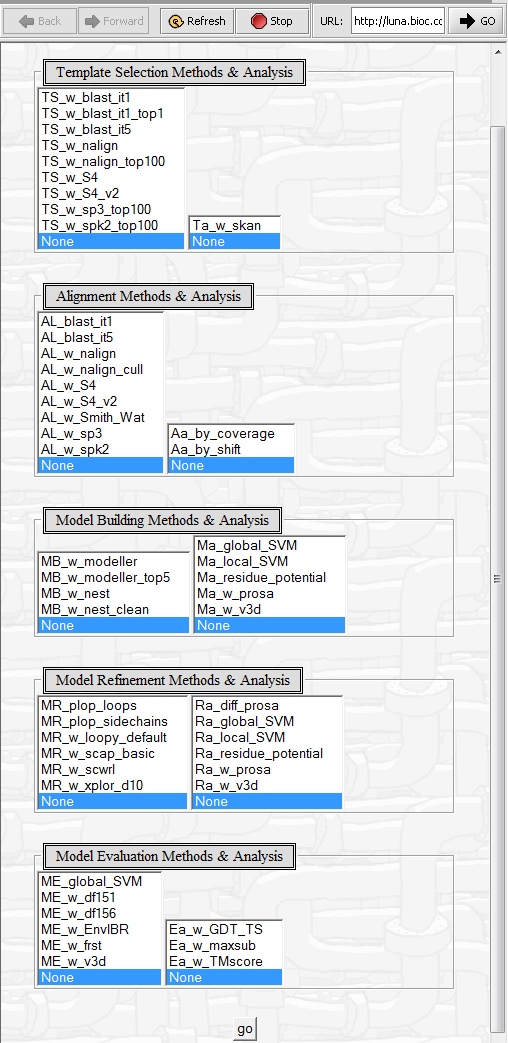
The Pudge analysis web page contains a number of configuration options.
Explanations of each available method can be obtained by clicking on its entry in the interface.
4. Select any desired methods - More than one per step can be chosen by use of the control key (Windows) while clicking in the menu. Once all selections have been made, click on Go to launch the job.
Results will be displayed in a web browser window, either your external browser or within geWorkbench (mostly no longer supported). From there, once can save result files in PDB format and load them into geWorkbench for viewing and further manipulation.
References
Norel R., Petrey D., and Honig B.: PUDGE: a flexible, interactive server for protein structure prediction. Nucleic Acids Research, 2010, Vol. 38, W550-W554 (Web Server issue). link to paper
Petrey D. and Honig B.: Protein structure prediction: inroads to biology. Mol Cell. 2005 Dec 22;20(6):811-9. link to paper