GeWorkbench Release 1.1
From Informatics
Contents |
Functionality Enhancements and Modifications
Marker and Array Set Combination Operations
Users can create new marker and array sets by applying intersection, union and exclusion operations on multiple selected sets. For a full description of the functionality, see Mantis report #992.
New version of caBIO
The Marker Annotations component has been updated to use the new version 4.0 of the caBIO API.
Server Side Computing
Full support for server side computing has been added (Dispatcher architecture) to facilitate the execution of long running jobs. The new architecture allows submitting an analysis request in one application session and retrieving the analysis results in another.
Workspace Saving Upon App Exit
The application now queries the user if they want to save their workspace before exiting.
Integration of Marker Annotation/caBIO Pathways Components
These two components have been combined into one per the requirements specified at:
http://wiki.c2b2.columbia.edu/mantis/view.php?id=1302
Not Yet Implemented
Support for Illumina Oligo chips
Data Format
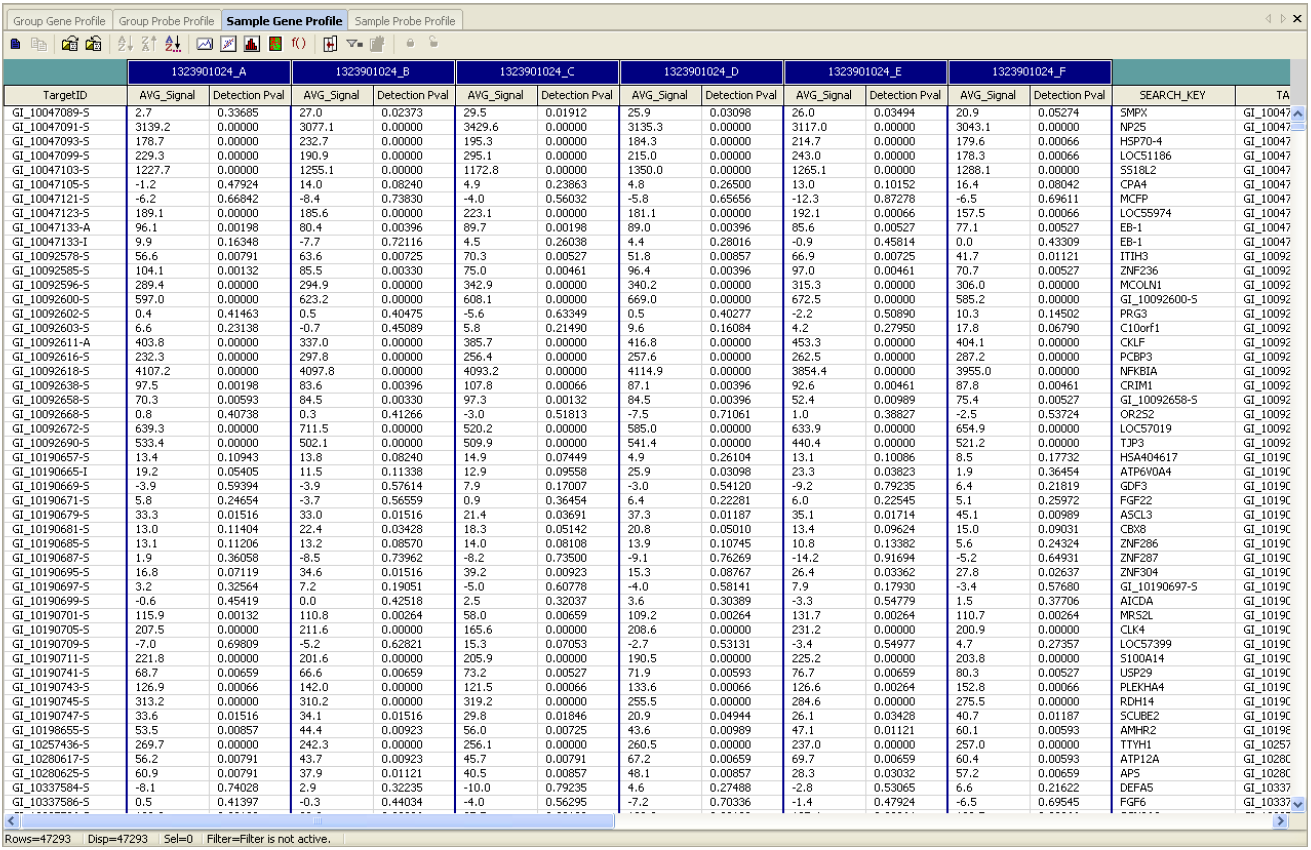
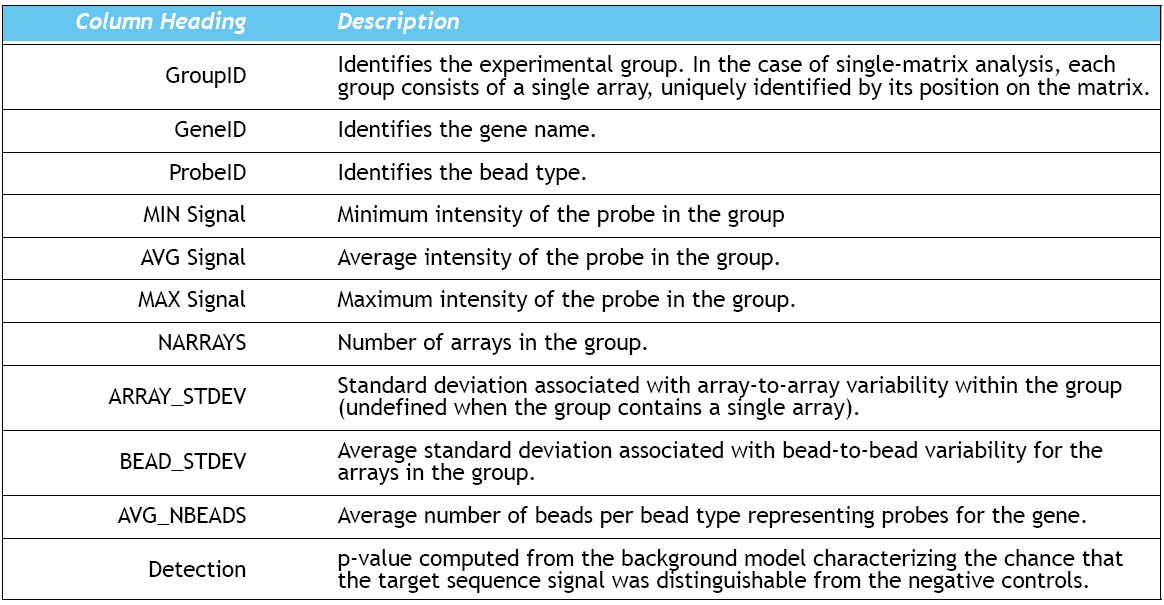
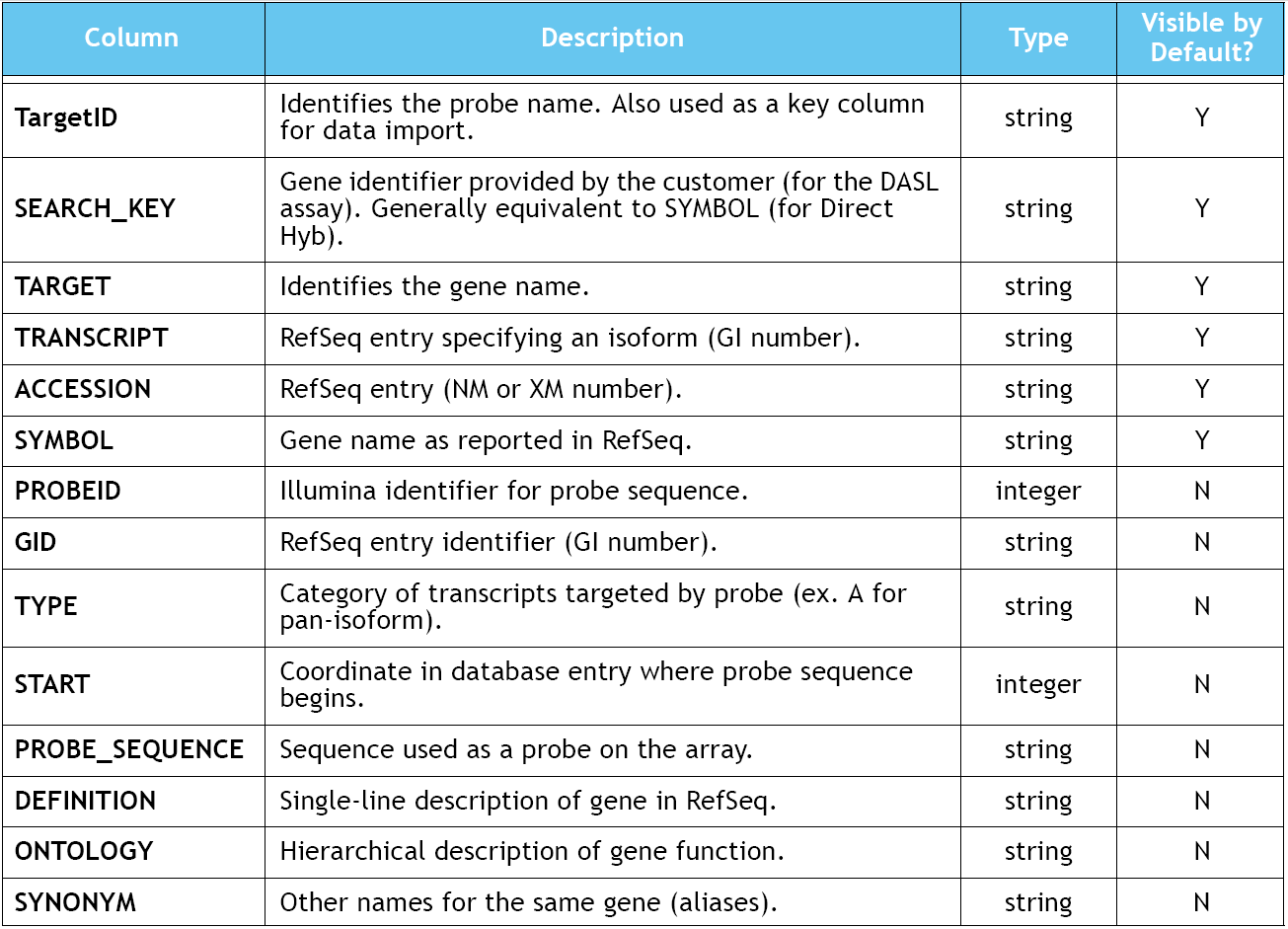
Format specification of the Illumina chips is as follows (Illumina raw data):
Source: BeadStudio Gene Expression Module: User Guide p.110 (Illumina, Inc. 2006)
Source: BeadStudio Gene Expression Module: User Guide p.111 (Illumina, Inc. 2006)
Source: Gene Expression on Sentrix Arrays: Direct Hybridization System Manual p.103 (Illumina, Inc. 2003)
Sample raw data file from Don Swan/Alex Yemelyanov
Correponding BioConductor File (for reference and convenience)
FAQ
- In the BeadStudio 2 Gene Expression Module, the MIN_Signal, AVG_Signal, and MAX_Signal columns all have the same value. What is wrong?
Nothing is wrong with the data. The MIN, AVG, and MAX columns only have different values when the grouping function is used to analyze multiple arrays or BeadChips. If you look at the NARRAYS column, the number 1 appears. This means that only 1 array was reported as a group in this analysis; therefore, the MIN, AVG, and MAX values are the same.