PUDGE Requirements
From Informatics
Overview
Background
In protein structure analysis, the crystal structure of a protein is sought after. As this can become experimentally expensive, a computational technique called protein prediction is used to identify the protein structure using protein sequence information. In protein prediction, there are methods that can be used either independently or together. These are:
- Ab initio - a simulation technique
- Homology Modeling
These techniques are used in collaboration within PUDGE. That is, PUDGE has a series of steps in the workflow which first utilize homology modeling and then Ab initio. These are detailed below.
Workflow
PUDGE has a number of steps in its workflow, each taking the output of the previous step as the input for the next step. Interacting with PUDGE for advanced users involves viewing and analyzing the output of one step and either rerunning the step with different input, or by modifying the output before it goes into the next step. The following is adapted from Donald Petrey's PUDGE manual:
http://wiki.c2b2.columbia.edu/honiglab/index.php/Structure_prediction_in_the_Honig_Lab
PIPELINE INPUT: A protein sequence. This is called the "target" sequence.
Stage 1 - Template selection
Used to predict the structure of a sequence with an unknown structure using homology, one must first find a protein or set of proteins which are likely to have a structure similar to the target. This is usually accomplished by finding proteins with experimentally determined structures which can be shown to have an evolutionary relationship to the target. Such proteins are usually referred to as "templates". In most homology modeling exercises, useful information can come from more than one template.
INPUT: A "target" protein sequence with an unknown structure OUTPUT: A list of "template" protein structures which have similar sequences to the target.
Stage 2 - Sequence-to-structure alignment (Change to Sequence Alignment)
(I do not think any structural alignment is being done here - KCS) To make a model, it is necessary to first identify the residues in the target sequence that are structurally equivalent to residues in the template structure. This pairwise correspondence of amino acids is called a "sequence-to-structure alignment", or simply an "alignment". It is not possible, in general, to generate the single "best" alignment without incorporating structural information. It is usually necessary to make many different alignments, especially for remote homologs (and even for close homologs in regions of low sequence identity, e.g. T0142 from CASP6).
INPUT: A list of protein structures which have similar sequences to the target OUTPUT: A series of residue-residue alignments of the templates to the target
Stage 3 - Model building
Given an alignment, an initial model is made by replacing the residues in the template with their corresponding residues in the target (based on the alignment). The backbone is generally kept fixed within secondary structure elements while the conformations of unconserved side-chains and insertions/deletions are predicted ab initio. Many models may be produced based on the available set of templates and alignments.
INPUT: A series of residue-residue alignments of the templates to the target OUTPUT: A set of model structures, which is not necessarily equal to the number of alignments
Stage 4 - Model refinement
While the assumption that structures will be similar if sequences are similar, it is usually the case that the target structure will be different from the template structure in one or several regions. Identifying and predicting the structure of these regions is called "model refinement" or simply "refinement". Refinement procedures may again produce multiple models.
INPUT: A series of model structures OUTPUT: A refined series of model structures
Stage 5 - Model evaluation
The steps described above may produce thousands of models. Choosing which is the best using some scoring function/effective energy function is called "model evaluation". A number of different methods are usually used starting with computationally inexpensive simplified scoring functions/statistical potentials to remove grossly incorrect models and subsequently applying more detailed physical-chemical energy functions to choose the "best" structure.
INPUT: A refined series of model structures OUTPUT: A set of structures, with a ranking
PIPELINE OUTPUT: A set of structures, with a ranking
Phase 1
PUDGE - geWorkbench Integration
Alpha Release
The alpha phase of this integration will substitute the existing web-form with geWorkbench as a user interface. The inputs on the interface are:
- protein sequence
- methods
This alpha release will be a basic, non-interactive integration. The sequence is submitted to PUDGE via the existing server functionality (explained below). PUDGE will then run with default parameters and return one PDB file if PUDGE can find any model to fit the sequence. This structure can be viewed using the existing functionality of geWorkbench (JMol).
Development Environment
All development are done from caldev.
The caGrid 1.0 is installed at caldev.
C:\Documents and Settings\xiaoqing\cagrid-1-0\
For external programs:
Globus ws-core 4.0.3 is installed at:
C:\Documents and Settings\xiaoqing\cagrid-1-0\external\ws-core-4.0.3
Tomcat 5.0.28 is installed at:
C:\Documents and Settings\xiaoqing\cagrid-1-0\external\jakarta-tomcat-5.0.28
The PUDGE code is created at:
C:\Documents and Settings\xiaoqing\PUDGE
Implementation
Set up PUDGE application server. To run standalone PUDGE, the following variables must be set up correctly as listed:
PUDGE=/razor/0/common/pudge PDB_DIRECTORY=/razor/0/databases/pdb TROLLTOP=/razor/0/common/pudge/dat/allh.top SUBMAT=/razor/0/common/pudge/dat/blosum62 JACKALDIR=/razor/0/common/pudge/dat/jackal.dir MOUNTDIR=/razor/0 JAVA_HOME=/nfs/apollo/1/server_data/www/pudge2/jdk1.5.0_05 PATH=$JAVA_HOME/bin:$PATH
Please note the cluster will use c shell and bash, so both .cshrc and .bashrc need change as the mentioned above. Currently the application server (Tomcat cat) is located at:
/nfs/apollo/1/server_data/www/pudge2/jakarta-tomcat-5.0.28
The user name is: pudgewb
Integration
When pressing submit on the UI, a sequence is submitted to the grid service (addwnld.) This service then creates corresponding fasta file and invokes a perl script to start the PUDGE pipeline. The script is located at:
/razor/0/common/pudge/scr/psub.pl.
The script will run the following command:
perl /razor/0/common/pudge/scr/psub.pl 2resub.cfg
After the perl script is called, the pipeline program will be invoked and then a working folder will be generated at:
/razor/0/pudge/common/pudge/wrk/P<rand4>/
2resub.cfg Format
<Option_Name><colon><tab><Selection_List>
where <Option_Name> is one of the following:
template_selection template_selection_analysis alignment alignment_analysis model_building model_building_analysis model_refinement model_refinement_analysis model_evaluation model_evaluation_analysis target_seq
<colon> and <tab> refer to the literal colon and tab characters
And <Selection_List> refers to a comma delimited (no spaces/tabs) list of options unique to each of: template_selection, template_selection_analysis, alignment, alignment_analysis, model_building, model_building_analysis, model_refinement, model_refinement_analysis, model_evaluation, model_evaluation_analysis
For target_seq, <Selection_List> is always "input_txt", identifying the target file mentioned above.
As discussed above, during the alpha phase PUDGE will use default method parameters to allow quick generation and retrieval of PDB files. These will be:
template_selection: test_template_selection template_selection_analysis: test_template_selection_analysis alignment: test_alignment_analysis model_building: test_model_building model_building_analysis: test_model_building_analysis model_refinement_analysis: test_model_refinement_analysis model_evaluation: test_model_evaluation model_evaluation_analysis: test_model_evaluation_analysis target_seq: input_txt
- At the end the pipeline program will dump the Protein Data Bank (PDB) file into a create a "models" folder under the working directory. The working folder will have a file titled "lock" when the job is running. The lock file will be removed and another file titled DONEALL.txt will show up when the job is done.
- The PDB file will be transferred back to geworkbench and displayed using JMol.
- The 2resub.cfg must be included in the tomcat/bin folder and all files in that folder must be executable.
The solution to the classloader issue.
THe standalone PUDGE client can connect with the service without any problem but when the code is plugged into geworkbench, an exception will be displayed:
java.lang.ClassCastException: org.apache.xerces.jaxp.DocumentBuilderFactoryImpl
To solve this problem, the j2ee.jar and arrayexpress.jar from the geworkbench core library need updated as follows: the old version of DocumentBuilderFactoryImpl needs to be removed from these two jar files. Fixed versions have been stored in CVS with the following names: j2eeWithoutDoc.jar and arrayexpressWithoutDoc.jar. These files are located in the geWorkbench lib and the caArrayMageom directory, respectively.
Finally, The parameters to run java need be set as following (you can do it either in build.xml or by entering it into the IDE (IDEA etc):
-Djavax.xml.parsers.SAXParserFactory=org.apache.xerces.jaxp.SAXParserFactoryImpl -Djavax.xml.parsers.DocumentBuilderFactory=org.apache.xerces.jaxp.DocumentBuilderFactoryImpl
The complete section in the build.xml file, after the addition of the two lines, looks like this:
<target name="run" depends="compile" description="Runs geWorkbench.">
<java fork="true" classname="org.geworkbench.engine.config.UILauncher">
<jvmarg value="-Xmx640M"/>
<jvmarg value="-Djava.library.path=lib"/>
<jvmarg value="-Djavax.xml.parsers.SAXParserFactory=org.apache.xerces.jaxp.SAXParserFactoryImpl"/>
<jvmarg value="-Djavax.xml.parsers.DocumentBuilderFactory=org.apache.xerces.jaxp.DocumentBuilderFactoryImpl"/>
<classpath refid="run.classpath"/>
</java>
</target>
Phase 2
Use Case
Primary actions
- User provides one or more sequences for which he/she wishes to model the 3-D structure.
- The sequences are loaded into geWorkbench as FASTA files.
- In a geWorkbench PUDGE interface, the user chooses methods for template selection, alignment, model building, refinement and evaluation. At each stage, one or more methods to analyze the stage results may also be chosen.
- Following the current PUDGE web interface practice, no parameters will be set manually. All analysis are prepackaged.
- (Alternatively, the selected FASTA file(s) could be sent to the server and the method selection process handed off to the PUDGE server itself via a web browser window. This would allow access to the latest improvements made by the PUDGE group.)
- The user submits the job.
- The user can monitor job progress:
- The list of chosen methods and analyses is displayed so that progress through them can be determined.
- is the job still running?
- what is the cumulative CPU time used?
- which stages have completed, or
- within a stage, which methods have completed (if multiple were chosen)?
- The user can examine intermediate and final results. The primary results are:
- Lists of alignments or structures and their P-values.
- Output from separate Analysis methods available at each stage - the results are graphical web pages.
- In addition, further visualizations are available for the primary alignments or structure files:
- Web page data visualizations generated by the "Show Results" program.
- Final results, both sequence alignments or 3-D structures, can be imported into geWorkbench. The existing web server provides a mechanism for concatenating the result files for export.
- Imported alignments or structures can be viewed in geWorkbench.
- For structure, the existing JMOL viewer can be used.
- For alignments, a new viewer may be needed - the alignment files are text.
- Complete or selected results should also be directly downloadable to the user's computer. The existing web server provides a mechanism for concatenating the result files for export.
- The user can resubmit a job from an intermediate step. For example, he/she may wish to choose a different modeling algorithm or analysis.
- Resubmission is only allowed for completed jobs.
- Intermediate resubmissions of the job run in the same directory structure under the same jobID.
- The user can rerun the entire job, e.g. if it has crashed and must be restarted.
Overview of the current web-based PUDGE
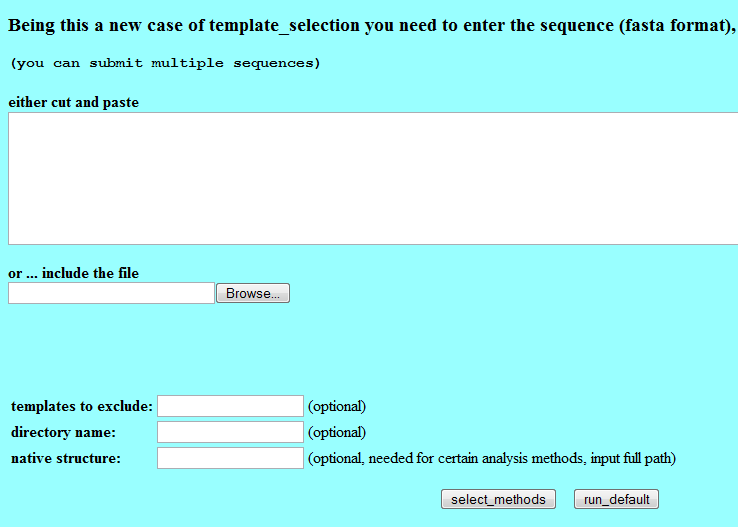
Sequence selection
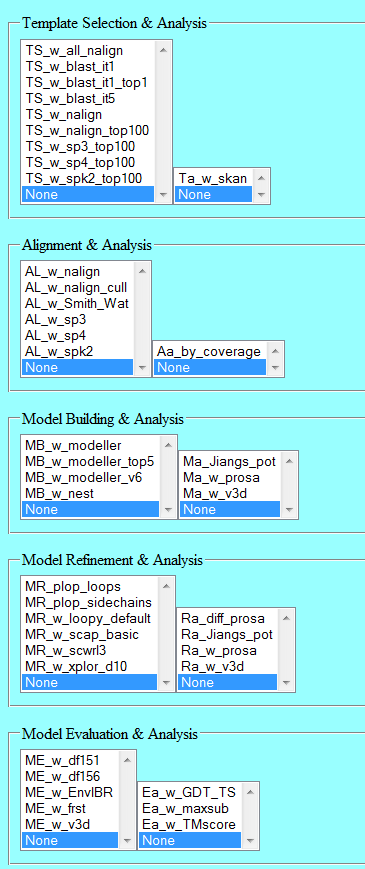
Method selection
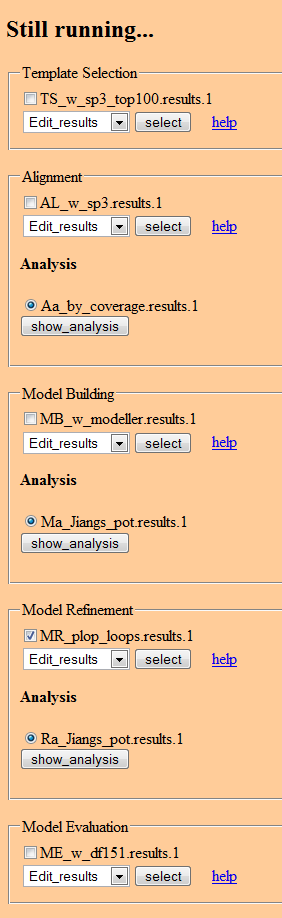
Job status page
Status page:
Details of current web-based PUDGE
Each pipeline stage result section has a pulldown menu. The menus vary slightly by stage. The contents are listed below by stage:
Template Selection
Top-level options are
- Edit_results - presents a list of sequences and alignment p-values
- Edit_sorted - presents a sorted list of sequences and alignment p-values
- Start_New - start a new job from the beginning, that is at the method selection screen.
- Re-Run - restart the job with the previous choices, overwriting any existing output files.
- Show_output - standard output from pipeline
- Show_errors - standard error from pipeline
Options - within the "Edit_results" and "Edit_sorted" menu choices, a menu with the following further choices is presented:
- save as edited - this is in fact a dead-end which is usable only from the server side, not in the web interface. It may be moved down in the list or dropped.
- resubmit - resubmit starting at following step (in this case, that would be alignment).
- results to TPR - outputs a file containing the names and p-values of all selected sequences, plus concatentated PDB coordinate models.
- obtain Annotation - (not working)
Alignment
Top-level options are
- Edit_results
- Edit_sorted
- Re-Run
- Show_output
- Show_errors
Options - within the "Edit_results" and "Edit_sorted" menu choices, a menu with the following further choices is presented:
- save as edited.
- resubmit
- results to TPR
- compareAlignments
- compareAlignments+Map
- download alignments
Model Building
Top-level options are
- Edit_results
- Edit_sorted
- Re-Run
- Show_output
- Show_errors
Options - within the "Edit_results" and "Edit_sorted" menu choices, a menu with the following further choices is presented:
- save as edited.
- resubmit
- results to TPR
- MapAnnotation_Mark-US
- download models
Model Refinement
Top-level options are
- Edit_results
- Edit_sorted
- Re-Run
- Show_output
- Show_errors
Options - within the "Edit_results" and "Edit_sorted" menu choices, a menu with the following further choices is presented:
- save as edited.
- resubmit
- results to TPR
- download models
Model Evaluation
Top-level options are
- Edit_results
- Edit_sorted
- Re-Run
- Show_output
- Show_errors
Options - within the "Edit_results" and "Edit_sorted" menu choices, a menu with the following further choices is presented:
- save as edited.
- resubmit
- results to TPR
- compareAlignments
- compareAlignments+Map
- obtainAnnotation
- download alignments
Web-based Pudge results displays (including "Show Results" program)
An overview of the result pages generated by the Show Results program is available:
http://wiki.c2b2.columbia.edu/honiglab_public/index.php/Software:Show_Results
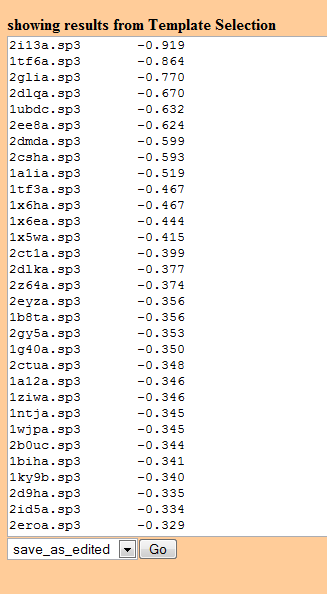
Text lists of sequence alignments or model structures and their p-values
The first menu item available for each pipeline section on the Results pages is "Edit Results". This presents an editable listing of sequences or model structures. If edited and saved, further visualizations will use the edited list. The original list can be obtained by returning to the results page and selecting "Edit Results" again.
Example of visualization of Sequence Alignments
- Multiple Sequence Alignments (MSAs) are generated using ?????
- The MSA is stored as a (JPEG)???? file.
- Also shown are features from the 3-D template such as cavities.
The Compare Alignments selection produces output such as shown here:
Example of Analysis of Sequence Alignments
If the Analysis option is chosen (Aa_by_coverage.results.1), a graphic web page is generated that itself has various options available via the menus on the left hand side:
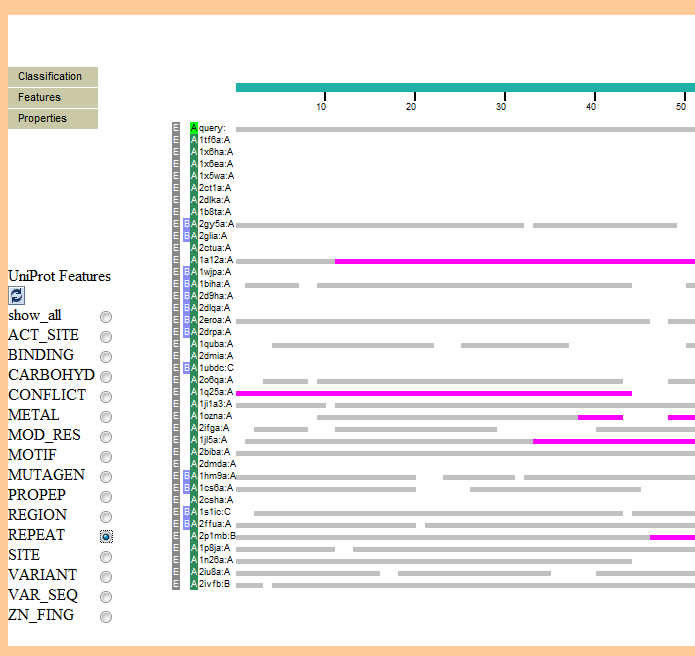
Here is the Features->UniProt submenu from that analysis:
Example of Analysis in the Model Building Section
An example of an Analysis using the "Jiang Potential" is shown:
Example of Analysis in the Model Refinement Section
An example of an Analysis using the "Jiang Potential" is shown:
Plan for full integration of Pudge into geWorkbench
Introductory comments on the proposed design
As shown in the screenshots in the previous section, the web-based Pudge provides a growing number of custom visualizations. Some of these are interactive, with further menus etc. The displays are under active development. This advanced visualization functionality cannot be quickly duplicated in geWorkbench, and doing so would server little purpose at present.
The proposed implementation of Pudge in geWorkbench is to use geWorkbench to build, launch, control and monitor jobs. The primary results can be displayed in geWorkbench, and sequence alignments and model PDB structures can be imported back into geWorkbench if desired.
It is proposed that the advanced visualizations be handled as at present via the Pudge server itself through a pop-up web browser window. The import of data back into geWorkbench would be performed directly through geWorkbench. No changes to the Pudge server are proposed, other than whatever changes might be needed to allow the geWorkbench-Pudge server access to its methods.
Sequence selection
The starting protein sequence(s) should be obtained from the Project Folders component. Note that this component shows sequences by FASTA file, not by individual sequence.
Pudge can handle multiple sequences in one input file (or pasted into the web interface).
Actions:
- The user loads an existing FASTA file using the "Open File(s)" option under the File menu.
- A new option to paste in FASTA sequences could be added under the File menu. If implemented, such a module should verify that the sequence is in the correct FASTA format.
- When a FASTA type object is selected in the Project Folders component, the PUDGE component should be materialized.
The PUDGE GUI component
A command module will be added to the geWorkbench commands area. It will contain the following tabs:
- Methods
- Execution
- Results
Visualizers
The following windows will be needed in the geWorkbench Visualization area.
- Text results - list of alignments and model structures with associated p-values.
- PDB 3D Structure files - visualize in JMOL, already present in geWorkbench
- Sequence Alignments - may need to develop a new viewer or find/develop implementing component.
PUDGE Method selection tab
Overview
The pipeline steps are:
- Template Selection
- Alignment
- Model Building
- Model Refinement
- Model Evaluation
Each stage also includes a separate list of one or more analysis methods that can be selected.
The current web-based Pudge uses a single web page with multiple-selection menus for each stage. This allows more than one method or analysis to be selected at any stage. It also allows the contents of the menus to be dynamically loaded, as new methods may be made available by the PUDGE group at any time. The nice thing about the current web layout is that all choices are visible at once. This is important because there are dependencies between stages.
Note that on the website each title also includes the word Analysis, e.g. "Template Selection & Analysis" but the repetition of "Analysis" is unnecessary in the context of geWorkbench.
Possible GUI designs:
- Implement a single page of dynamically generated list boxes just as PUDGE does now. This is simple and keeps geWorkbench consistent with the original PUDGE.
- A JTree like component as used in the geWorkbench Markers/Arrays/Phenotypes component. Rather than independent menus, there would be a single tree with branches.
- The first level would be tree nodes representing each stage of the pipeline
- In the second level would be labeled checkboxes representing each available method. Analysis methods would also be included. The analysis methods could be differentiated by a spaceholding label or by color etc.
- The user would select all desired methods and analyses.
- There should be a "Show All" methods button. This would expand the tree to show all entries.
- A more complex setup where only pulldown menus for each stage are shown. There could be a menu for each stage for pipeline methods and one for analysis methods. A list box would be used to receive and represent the chosen pipeline methods, with each stage clearly delimited by a label. A second list box next to the first would hold analysis selections. The list would be properly managed so that chosen items are inserted in the proper pipeline order, regardless of the order they are selected.
- The selected menu item is added to the appropriate section of the list box.
- Items could be deleted from the list box.
(I currently like the JTree design (2) best).
A key design constraint is that, if on the Results tab, when viewing results for a particular stage, the user chooses resubmission, only the configuration menus for the subsequent steps of job submission are presented - that is, the user is guided through the appropriate sequence of available steps. For example, if the user is in the Alignments section and chooses to resubmit the job, the user is returned to the job configuration menu showing only steps from Model Building on.
Method Descriptions (Hover Text) and Dependencies
Note that in the web version, each method has hover text that explains the method and any dependencies. This should be duplicated in geWorkbench. Because this content is dynamic, may be augmented or corrected at any time, and is needed in real time to make choices due to dependencies between stages, it is not recommended that it be placed in the online help.
- The geWorkbench version could provide hover text as done in the web version, or
- the methods/analysis definitions could be displayed in a text box to the right of the menus.
- An unresolved issue - how to keep the descriptive text in geWorkbench current with that in the web version?
Note - the method menu item selections do not respect known dependencies. That is, one is allowed to choose any methods, and if there is a mismatch between two stages, the requested methods will fail but the job will keep running. It is regarded by the Pudge developers as too difficult to implement a full matrix of dependencies because new methods are always being added. It is up to the user to understand the choices he or she is making.
Each pipeline stage (section of the submission page) should offer the methods shown in the corresponding section of the online Pudge, except any for which we do not have a valid license or right to use (check on Modeler for example).
Execution Tab
The execution tab should have the following:
- Submit Job (Button) - send pipeline configuration file to the Pudge server.
- Stop Job (Button) - send a terminate signal to the Pudge server (if possible).
- Restart Job (Button) - Start the job again from the beginning, without alteration. Note that if this is implemented, this option can be removed from the method menus.
- This is meant to be used if a job has died. If it is still running, a warning should be displayed that the current job should be stopped first.
- Update job status (Button) - retrieve the current job status from the Pudge server and display it.
- Job Status (Text box) - displays information about current job status. See screenshot of existing PUDGE job status above.
- Test Pipeline (Button)- (Optional) Execute a simple, pre-tested script that quickly exercises each stage of the pipeline and is known to work.
- JobID (TextBox) - Upon start of execution, a jobID object should be placed in the Project Folders component, and the alphanumeric jobID should be placed in a text box in the execution tab. This jobID should be compatible with the existing Pudge web interface, such that results generated using geWorkbench should also be visible in the web interface.
- Update Methods (Button)- Should there be a method to update the menu selections against the method database maintained on the Pudge server? (or automatic?)
Web-based PUDGE includes three text fields on the sequence entry page for modifying aspects of the job:
- Templates to exclude - a text field allows a PDB structure designator to be entered that the user wishes to exclude from consideration.
- Directory name - this allows a folder to be specified for e.g. results on kinases. Job results will then be placed in this subfolder on the server. These folders can be viewed and all available results browsed - this provides a means to keep track of one's work. Otherwise, jobs are saved only by jobID.
- Native structure - Can be used by Skan analysis.
Note - should we implement a function to retrieve job status or results based on a Job-ID entered as text?
Results Tab - GUI Layout
Pudge currently displays each available result set and allows one or more result set at each stage to be selected and then acted on by a menu choice, e.g. Edit, Edit sorted etc. This display directly represents the state of job progress.
Choice 1 - just like Pudge
This page could be reimplemented in geWorkbench with similar functionality.
- The available result sets should be listed in the results tab with check-boxes, separated by pipeline stage.
- The menu of available actions for each stage should be presented. Note that this menu is the same for each stage save the first, "Template Selection".
- Since we only want to select/edit results from one stage at a time, a separate "Select" button for each stage seems necessary, as in the web interface (better ideas welcomed here!!).
- When one or more result sets is selected, the contents should be displayed in the visualization area in an editable text box.
- When more than one result set (check-box) is selected from a given stage, all selected result sets are merged.
- A link to the web version of Pudge should be provided for access to the advanced visualizations (perhaps a "Go to Pudge Web Visualizations" button).
- When pushed, if possible we should start a Pudge web browser window which will be in the same state as the geWorkbench window - that is, it will show the list of alignments/models, and the advanced visualizations can be chosen from the web page.
- A Retrieve Results button should be provided for each stage - retrieve the alignments or structures for a given stage and add it to the Project (see discussion below). (Caution - depending on the details of the run a very large number of alignment or structure files might be generated).
Choice 2 - Alternative designs to consider
- Rather than display all results outright, we could use a tree design (JTree) as in the Markers/Arrays/Phenotypes selection comoponents.
- The first level objects would be the pipeline stages, represented by exclusive radio buttons. Checking the radio button would configure the context specific menu for that stage.
- The second level objects would be checkboxes representing individual results. One or more boxes could be checked within each result set.
- A pulldown menu of context specific choices would be offered, dependent on the active radio button. The first item should be a null item, like "make selection" or "Alignment Action", "Model Action" etc....
- Making a selection in the menu would directly execute the action.
- There should be an "expand all" button so that all results can be displayed.
Results Tab - Operations
The primary output of each stage of Pudge is a list of alignments/structures with associated p-values. The secondary output of each stage of PUDGE is a set of sequence alignments (steps 1 &2) , structures (steps 3-5), or analysis files (any step). Additional visualizations are available (see description above).
The web-based version of Pudge allows intermediate results to be inspected as they become available.
In the web interface, the results are displayed in a linear fashion, not in a nested fashion. All template selections are displayed first, then all alignments etc. If multiple methods are used at one step, then at the end of the next step, for each method used, there will be a numbered file. The numbers are presumably assigned in the order that the methods are listed in the previous step. The set of methods used to generate any particular result can be determined by inspection of file names but is not directly indicated. (Note - we should store a list of steps used in Dataset History).
For simplicity geWorkbench should probably adopt the same, non-hierarchical result display for this phase of development.
Review of operations on Results
As already noted above in the description of Web Pudge, several operations are available on the results from each stage. The options available in the first stage, Template Selection, are:
- Edit_results - presents a list of sequences and alignment p-values
- Edit_sorted - presents a sorted list of sequences and alignment p-values
- Start_New - start a new job from the beginning, that is at the method selection screen.
- Re-Run - restart the job with the previous choices, overwriting any existing output files.
- Show_output - standard output from pipeline
- Show_errors - standard error from pipeline
Proposal for working with results in geWorkbench:
- Edit_results - This operation should copy the selected results to an Edit window.
- Edit_sorted - This operation should should copy the selected results (sorted) to an Edit window.
- Start_New - (Omit) There is no need for this function on the Results tab menus. It can be performed directly from the Methods tab.
- Re-Run - (Optional) This would re-launch the job on the server, presumably without regenerating the submission script.
- Show_output - display the STDOUT from the process in the edit window.
- Show_errors - display the STDERR from the process in the edit window.
Review of working with edited results
Once results have been copied to the Edit window, Web Pudge allows several operations on them, for example:
- save as edited - this is in fact a dead-end which is usable only from the server side, not in the web interface. It may be moved down in the list or dropped.
- resubmit - resubmit starting at following step (in this case, that would be alignment).
- results to TPR - outputs a file containing the names and p-values of all selected sequences, plus concatentated PDB coordinate models.
- obtain Annotation - (not working)
Proposal for working with edited results in geWorkbench:
- save as edited - Omit.
- resubmit - implement as below.
- results to TPR - Implement by saving the generated file as a text file in the Project Folders component.
- obtain Annotation - Omit.
Resubmission - Web Pudge allows results from a job to be resubmitted from an intermediate stage. The resubmission is done from the editable list of alignments/structures. If desired a subset of the results from a stage can be selected and submitted to the next stage of processing. In principle, one could even run one stage at a time and inspect the results before choosing results and methods for the next stage. One can also rerun the pipeline from an intermediate point with different methods. (Note - you cannot resubmit a job if the original is still running - the resubmit feature is meant as a way to modify and rerun already completed jobs).
If the resubmission option is implemented in geWorkbench, activating it should take one back to the Methods tab, with only the appropriate (following stages) methods available. The menu selections should be put into the state there were in in the originally submitted job. Note - this would require remembering the state of the original or last job submission of the current job. (Check if Pudge does this).
Pudge integration with the Project Folders
- The parent node will be a sequence file in the Project Folders component.
- Each run of Pudge should result in a node with name "Pudge_JobID" placed in the Project Folders component as a child of the parent sequence node. The appropriate value of JobID for that run should be used.
A Pudge run should appear as an object under its parent FASTA file. If there are multiple sequences in the input FASTA file, each separate run should be a unique node under the parent multiple sequence FASTA file.
Project Folders
FASTA file (one or more up to N sequences)
|
PudgeJobObjectID#1
|
PudgeJobObjectID#2
|
|
PudgeJobObjectID#N
The PudgeJobObject stored in the Project Folders component should contain or point to an updatable list of currently available result sets from that particular job. Each result set is the result of a particular method run in a particular stage (result files plus their p-values). As new result sets become available either the object should be regenerated or the new results appended. The primary results are just text files so a text file viewer should be supplied.
Open Questions
- Under each PudgeJobOBjectID#N could appear imported alignment or PDB structure files. Note that the Pudge server supplies e.g. all requested PDB structures concatenated into one file. We must investigate if we want to maintain the data in this kind of format, as we do for multiple FASTA files, or unpack them and represent them individually, perhaps in subfolders if possible (recommended).
- Note that if alignments or structure files are imported into the Project Folder there is no guarantee that they remain current - e.g. subsequent re-runs of the job could add more results to a given stage and require re-evaluation. For now this must be left to the user to manage.
- The main unresolved question regards long term data storage and availability. The intermediate and final results of a Pudge modeling run reside on the server.
- Do we guarantee long-term storage, or
- should we provide the ability to generate a ZIP file of the entire run directory?
- Should the edit window for primary text results (names of sequence templates/alignments) be a part of the Results tab or a separate visualization window. Note that it may implement the "Resubmission" function, which will take the user back to the Method Selection tab of the Pudge component (in the Analysis section of geWorkbench).