Difference between revisions of "Home"
(→Brief Overview of Changes in geWorkbench 2.2.1) |
|||
| Line 33: | Line 33: | ||
==Brief Overview of Changes in geWorkbench 2.2.1== | ==Brief Overview of Changes in geWorkbench 2.2.1== | ||
| − | + | This release augments the recently released version 2.2.0, and includes more than 90 additional enhancements and bug fixes, with many focused on network import, display and export, sequence retrieval, and pattern discovery. It also corrects two omissions from release 2.2.0. | |
| − | |||
| − | discovery. | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | 2.2.0 | ||
| − | |||
| − | |||
| − | |||
| + | * Alternate file viewer for large networks that may be too large to view in Cytoscape. | ||
| + | * Improvements to display of probeset-level networks. | ||
| + | * Networks can now be imported either from ARACNe adjacency matrix files or from SIF files. Networks can be represented by gene symbols, Entrez IDs, probeset names, or other identifiers. | ||
| + | * Adds support for use of alternate ontology files (e.g. from the GO website) for gene ontology analysis. | ||
| + | * Sequence retrieval for DNA sequences is now based on refSeq records from the UCSC refGene table, and is available for all organisms with genomes supported by UCSC. | ||
| + | * The new "Fold Change" analysis component, omitted from release 2.2.0, is included. | ||
| + | * A feature to overlay a t-test result onto a Cytoscape network, not functional as released in version 2.2.0, now works correctly. | ||
| + | * Bonferroni correction added to the ARACNe GUI. | ||
| + | For a full list of changes, see [[GeWorkbench_2.2.1_Changes.txt| Changes]]. | ||
| + | |||
| + | geWorkbench v2.2.1 can be downloaded from https://gforge.nci.nih.gov/frs/?group_id=78. Full installation instructions are available there in the Release Notes, and also on the geWorkbench website at http://www.geworkbench.org. In-depth documentation is also available in the geWorkbench [[Tutorials|Tutorials]]. | ||
==Previous Releases== | ==Previous Releases== | ||
See the [[PreviousReleases | Previous Releases]] page for full details. | See the [[PreviousReleases | Previous Releases]] page for full details. | ||
Revision as of 14:33, 15 August 2011
Contents
Quick Start
Please see the Quick Start guide to geWorkbench to see how to get started using geWorkbench right away. We are continuing to develop new material for this guide.
Overview
Welcome to geWorkbench. The current version is 2.2.1, released July 29th, 2011.
See the list of the most important fixes, changes and enhancements. This release was entirely devoted to improvements of existing components. More than 90 issues were addressed. The most important are summarized below.
The latest Release Notes and downloads can be obtained from https://gforge.nci.nih.gov/frs/?group_id=78. Installation instructions can be found on the Download and Installation page of this Wiki.
geWorkbench (genomics Workbench) is a Java-based open-source platform for integrated genomics. Using a component architecture it allows individually developed plug-ins to be configured into complex bioinformatic applications. At present there are more than 70 available plug-ins supporting the visualization and analysis of gene expression and sequence data. Example use cases include:
- loading data from local or remote data sources.
- visualizing gene expression, molecular interaction networks, protein sequence and protein structure data in a variety of ways.
- providing access to client- and server-side computational analysis tools such as t-test analysis, hierarchical clustering, self organizing maps, regulatory neworks reconstruction, BLAST searches, pattern/motif discovery, etc.
- validating computational hypothesis through the integration of gene and pathway annotation information from curated sources as well as through Gene Ontology enrichment analysis.
geWorkbench is the Bioinformatics platform of MAGNet, the National Center for the Multi-scale Analysis of Genomic and Cellular Networks (one of the 7 National Centers for Biomedial Computing funded through the NIH Roadmap). Additionally, geWorkbench is supported by caBIG®, NCI's cancer Biomedical Informatics Grid initiative.
End-user and developer support for geWorkbench is provided through the caBIG® Molecular Analysis Tools Knowledge Center, a component of the caBIG® Enterprise Support Network.
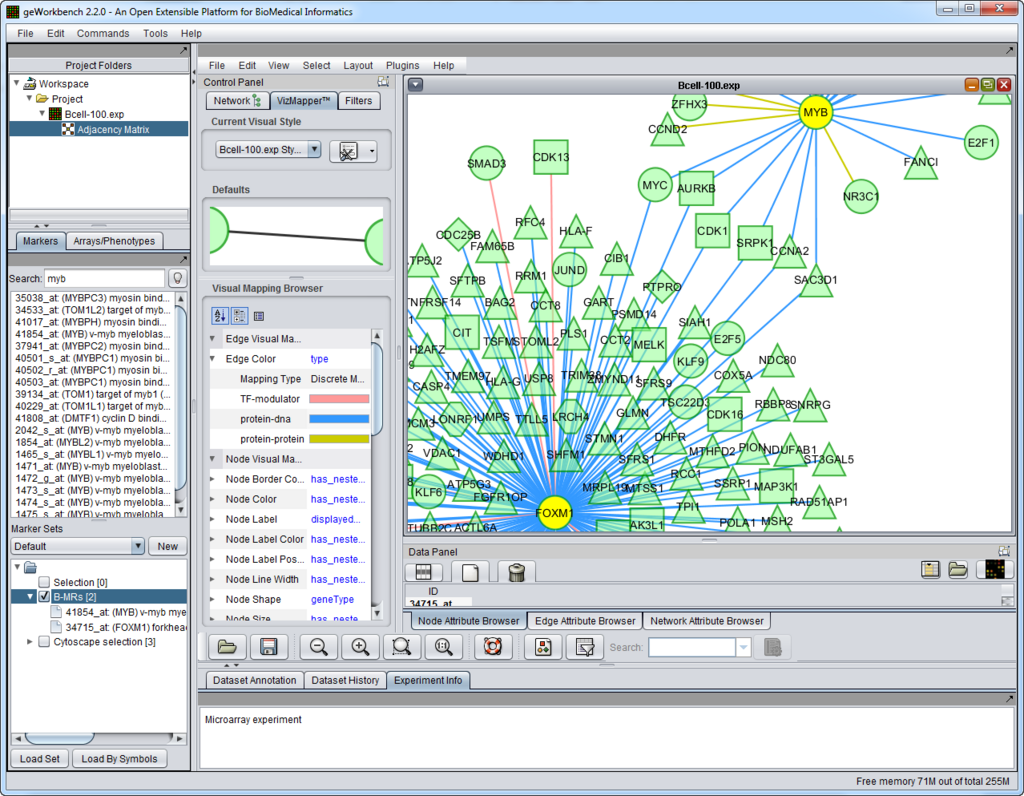
Graphical User Interface
The figure above shows the full geWorkbench graphical user interface. The Cellular Network Knowledge Base B-cell interactome was queried for interactions of two master regulator genes, and the result displayed in Cytoscape.
Brief Overview of Changes in geWorkbench 2.2.1
This release augments the recently released version 2.2.0, and includes more than 90 additional enhancements and bug fixes, with many focused on network import, display and export, sequence retrieval, and pattern discovery. It also corrects two omissions from release 2.2.0.
- Alternate file viewer for large networks that may be too large to view in Cytoscape.
- Improvements to display of probeset-level networks.
- Networks can now be imported either from ARACNe adjacency matrix files or from SIF files. Networks can be represented by gene symbols, Entrez IDs, probeset names, or other identifiers.
- Adds support for use of alternate ontology files (e.g. from the GO website) for gene ontology analysis.
- Sequence retrieval for DNA sequences is now based on refSeq records from the UCSC refGene table, and is available for all organisms with genomes supported by UCSC.
- The new "Fold Change" analysis component, omitted from release 2.2.0, is included.
- A feature to overlay a t-test result onto a Cytoscape network, not functional as released in version 2.2.0, now works correctly.
- Bonferroni correction added to the ARACNe GUI.
For a full list of changes, see Changes.
geWorkbench v2.2.1 can be downloaded from https://gforge.nci.nih.gov/frs/?group_id=78. Full installation instructions are available there in the Release Notes, and also on the geWorkbench website at http://www.geworkbench.org. In-depth documentation is also available in the geWorkbench Tutorials.
Previous Releases
See the Previous Releases page for full details.