Difference between revisions of "Screenshots"
| Line 63: | Line 63: | ||
Compare multiple markers or arrays with the standard Scatter Plot analysis. | Compare multiple markers or arrays with the standard Scatter Plot analysis. | ||
| − | Array vs Array | + | ====Array vs Array==== |
[[Image:GeWB_Scatter_Plot_array_vs_array.png]] | [[Image:GeWB_Scatter_Plot_array_vs_array.png]] | ||
| − | Marker vs Marker | + | ====Marker vs Marker==== |
[[Image:GeWB_Scatter_Plot_marker_vs_marker.png]] | [[Image:GeWB_Scatter_Plot_marker_vs_marker.png]] | ||
| Line 73: | Line 73: | ||
---- | ---- | ||
| − | === | + | ===T Test=== |
| − | + | ====Volcano Plot==== | |
| + | A t-test result display on a "volcano plot": Log significance vs log fold change. | ||
| − | [[Image: | + | [[Image:T_t-test_volcano_BCELL_webm_qldm.png]] |
| + | |||
| + | ====Color Mosaic==== | ||
| + | The t-test result can also be displayed as a color mosaic. | ||
| + | |||
| + | [[Image:T_t-test_colormosaic_BCELL_webm_qldm.png]] | ||
---- | ---- | ||
| + | === Hierarchical Clustering Dendrogram === | ||
| + | A Dendrogram displays the results of the Hierarchical clustering analysis. | ||
| − | + | [[Image:T_HC_Dendrogram_display.png]] | |
| + | ---- | ||
| + | === SOM Clustering === | ||
| + | Self Ordered Map clustering results are displayed as series of expression profiles corresponding to discovered groupings. | ||
| + | [[Image:T_SOM_result.png]] | ||
| + | ---- | ||
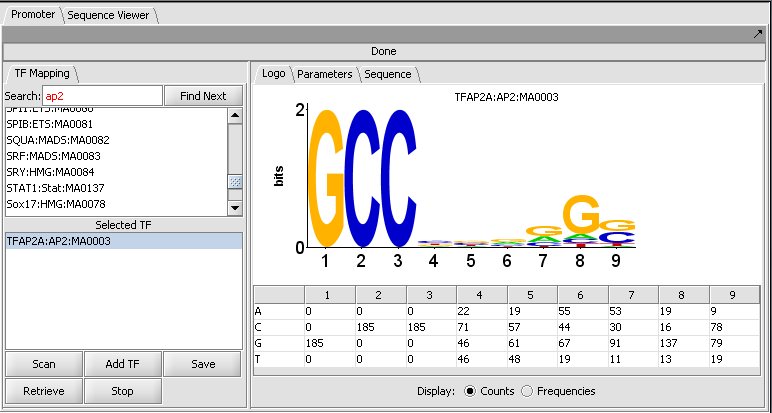
| + | ===Promoter Panel=== | ||
| + | Individual motifs from the JASPAR Transcription Factor Binding Profile Database can be scanned against loaded genomic sequences. | ||
| + | ====Motif selection and Logo display==== | ||
| − | + | [[Image:T_Promoter_CDH2_AP2_2000updn_setup_logo.png]] | |
| − | |||
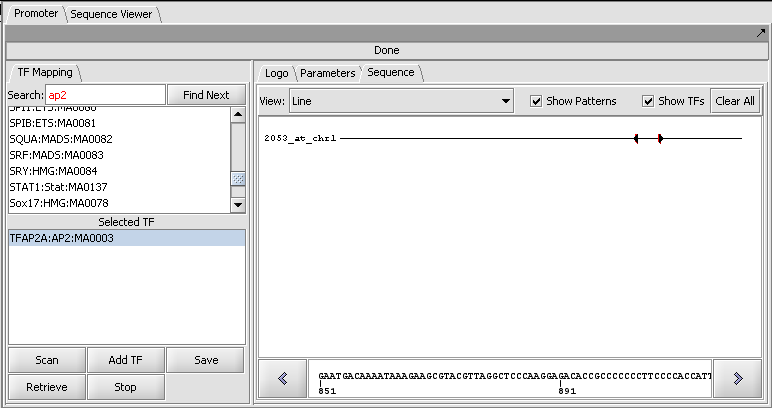
| + | ====Result of a scan against a single sequence==== | ||
| − | [[Image: | + | [[Image:T_Promoter_CDH2_AP2_2000updn_scan.png]] |
| − | |||
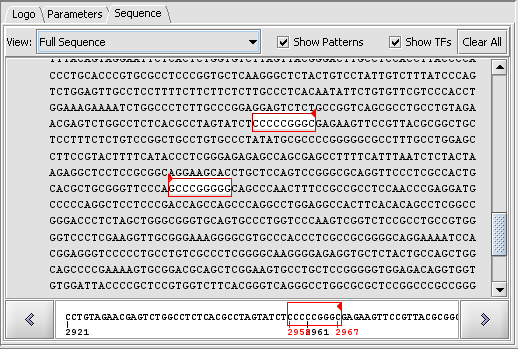
| − | === | + | ====Sequence-level display of match==== |
| − | |||
| + | [[Image:T_Promoter_CDH2_AP2_2000updn_scan_fullseq.png]] | ||
| − | |||
---- | ---- | ||
| + | |||
| + | |||
| + | == geWorkbench screenshots (previous)== | ||
| Line 124: | Line 141: | ||
---- | ---- | ||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
| − | |||
=== BLAST Queries === | === BLAST Queries === | ||
Revision as of 16:44, 9 February 2010
Contents
geWorkbench screenshots (Updated)
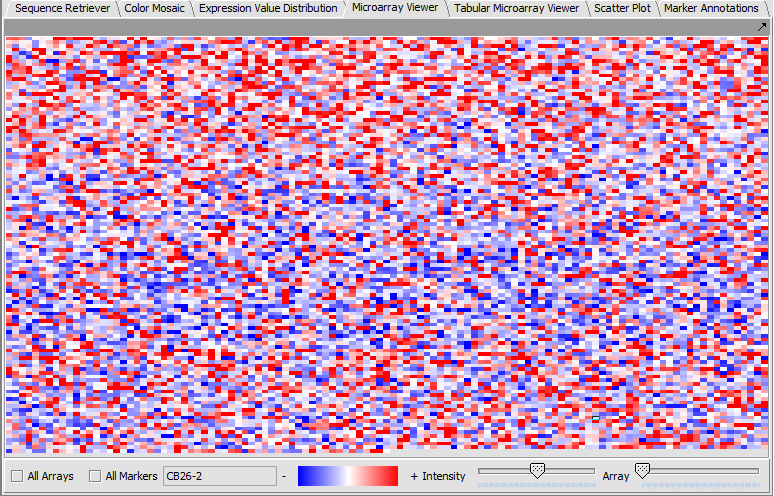
Microarray Viewer
The Microarray Viewer displaying marker values for selected array.
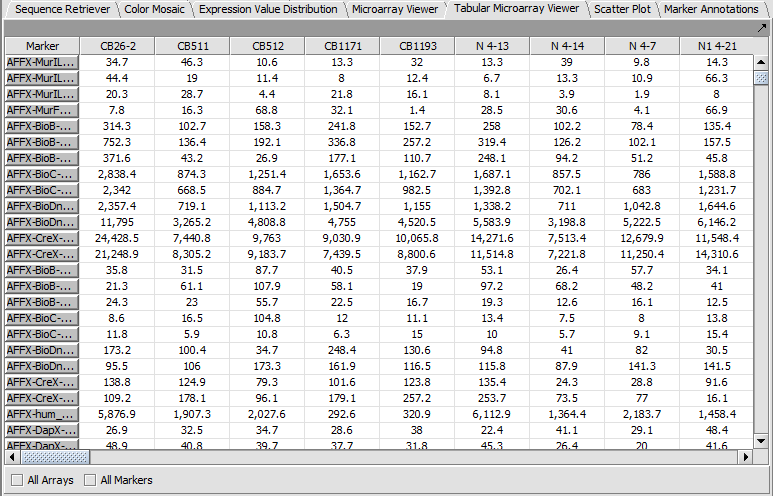
Tabular Microarray Viewer
The Tabular Microarray Viewer displays expression values in spreadsheet format.
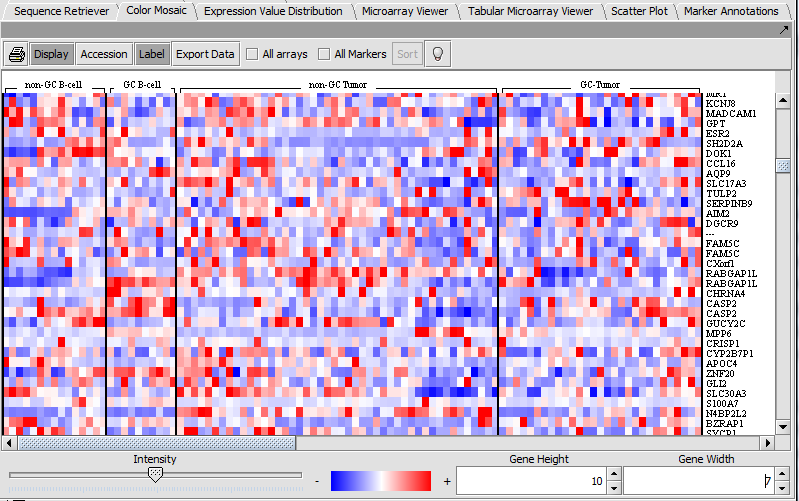
Color Mosaic
The Color Mosaic component displaying selected arrays, group designation and marker names.
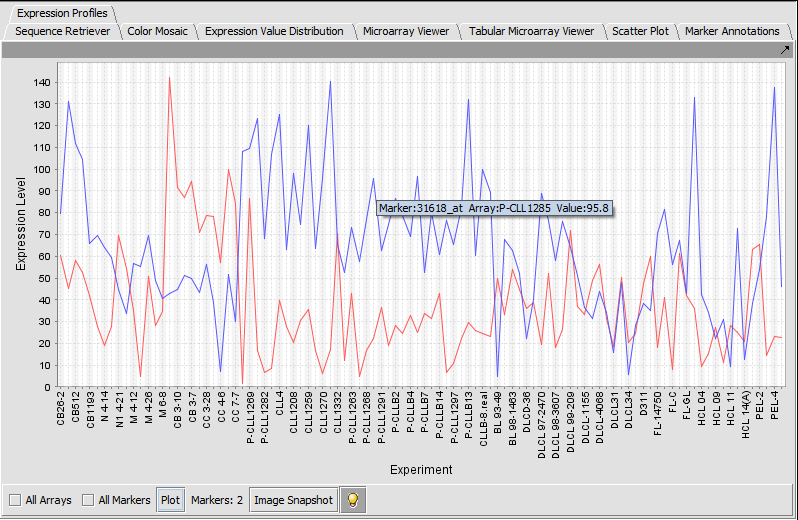
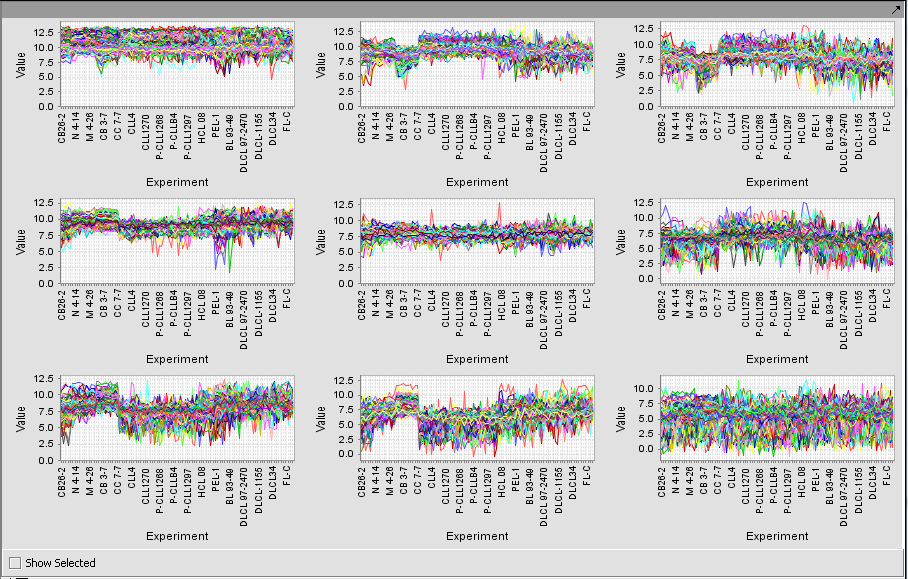
Expression Profile
Expression Profile plotting values for selected markers and arrays.
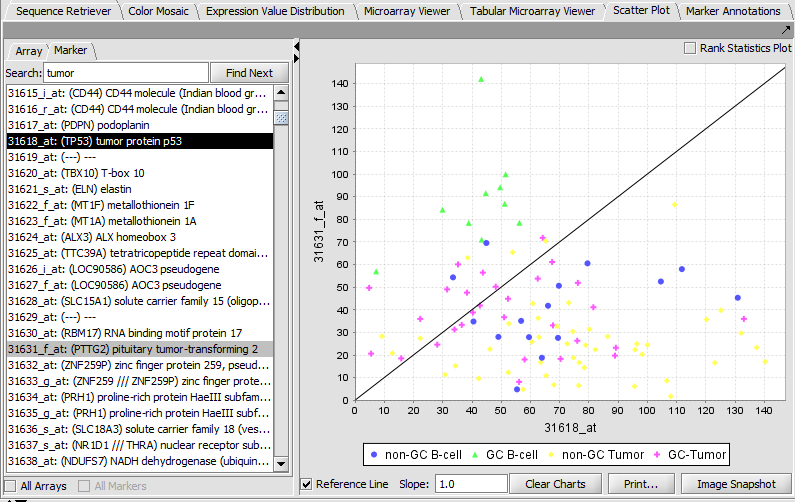
TP53 (31618) and PTTG2 (31631_f_at)
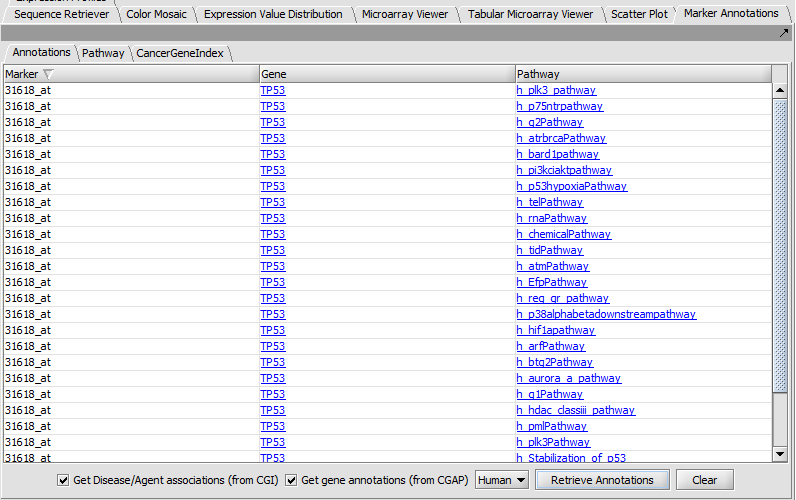
Marker Annotations
Retrieve and display gene and pathway information from CGAP and Cancer Gene Index (CGI).
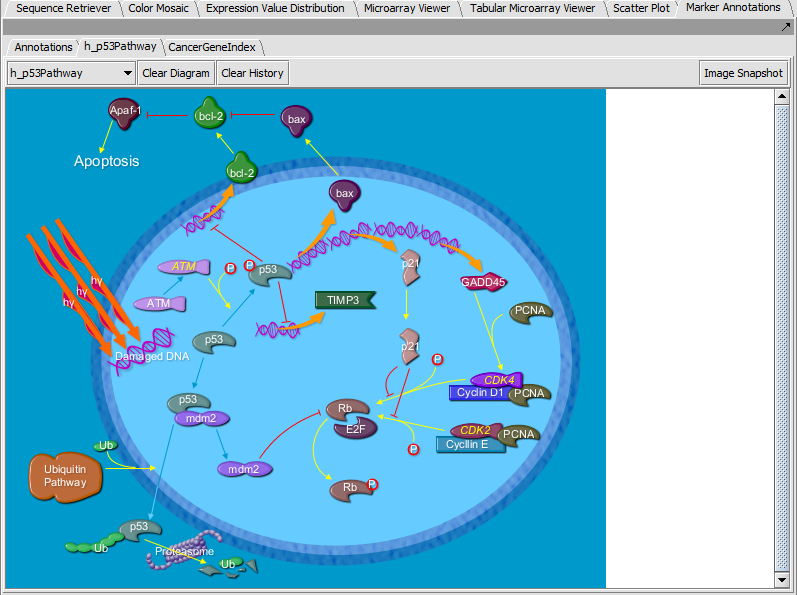
Marker Annotations - BioCarta Pathways
Displays BioCarta images retrieved from caBIO.
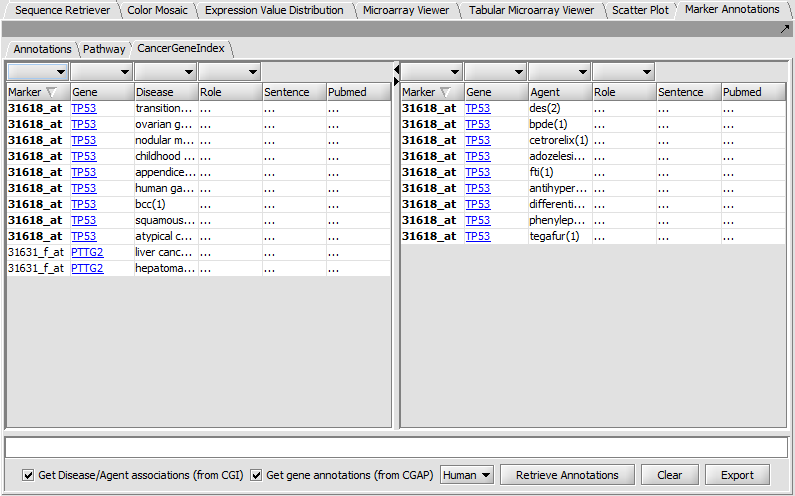
Marker Annotations - Cancer Gene Index
Displays literature citations from the Cancer Gene Index project.
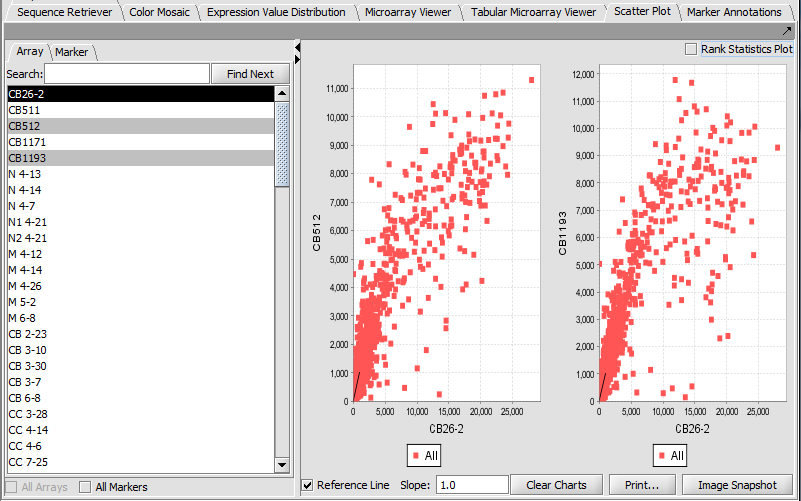
Scatter Plot
Compare multiple markers or arrays with the standard Scatter Plot analysis.
Array vs Array
Marker vs Marker
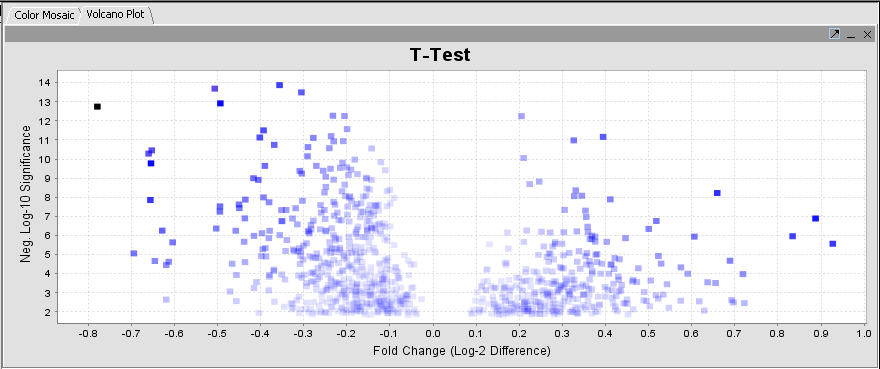
T Test
Volcano Plot
A t-test result display on a "volcano plot": Log significance vs log fold change.
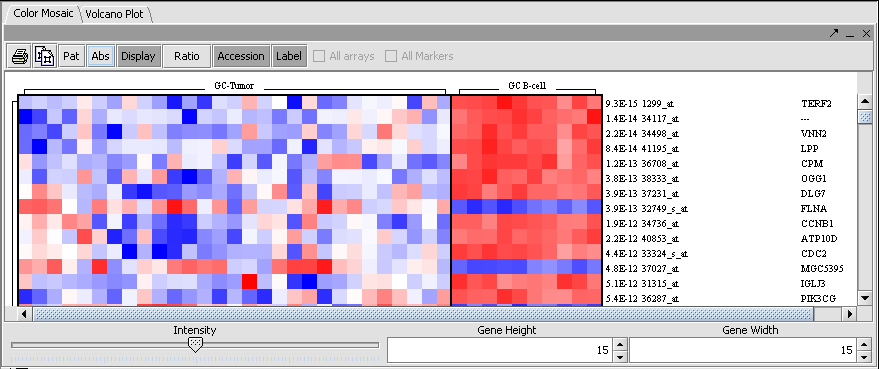
Color Mosaic
The t-test result can also be displayed as a color mosaic.
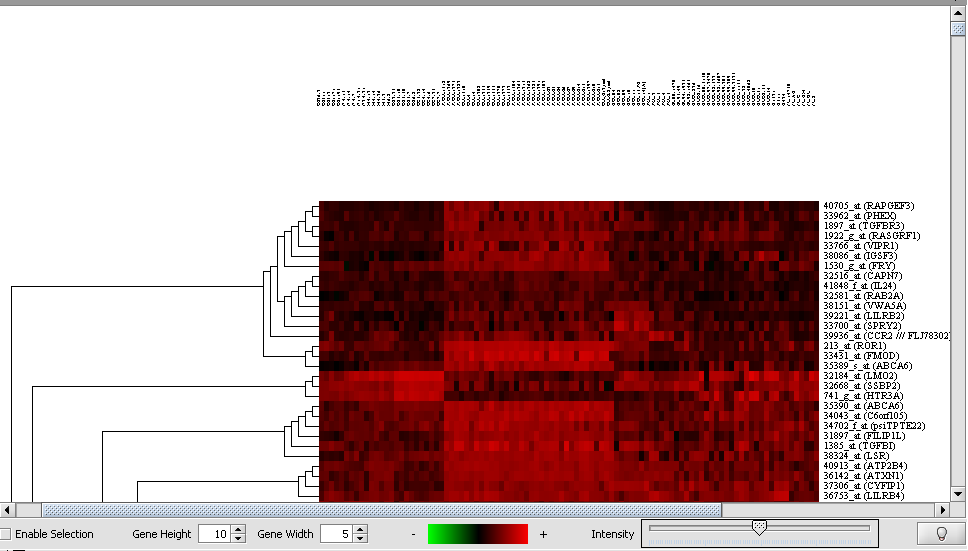
Hierarchical Clustering Dendrogram
A Dendrogram displays the results of the Hierarchical clustering analysis.
SOM Clustering
Self Ordered Map clustering results are displayed as series of expression profiles corresponding to discovered groupings.
Promoter Panel
Individual motifs from the JASPAR Transcription Factor Binding Profile Database can be scanned against loaded genomic sequences.
Motif selection and Logo display
Result of a scan against a single sequence
Sequence-level display of match
geWorkbench screenshots (previous)
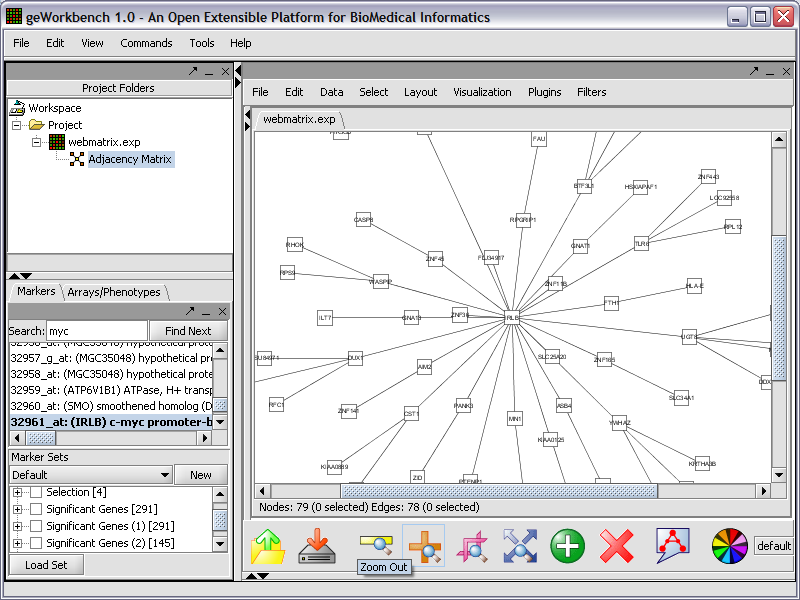
Cytoscape
Use the Cytoscape network visualization to view the reverse engineered regulatory network.
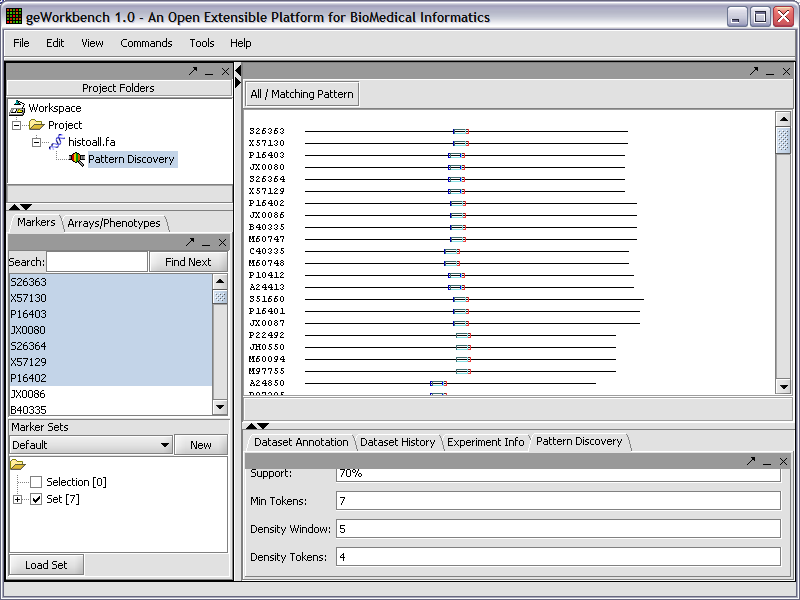
Pattern Discovery
Use the SPLASH algorithm to discover sparse amino or nucleic acid patterns in a loaded sequence.
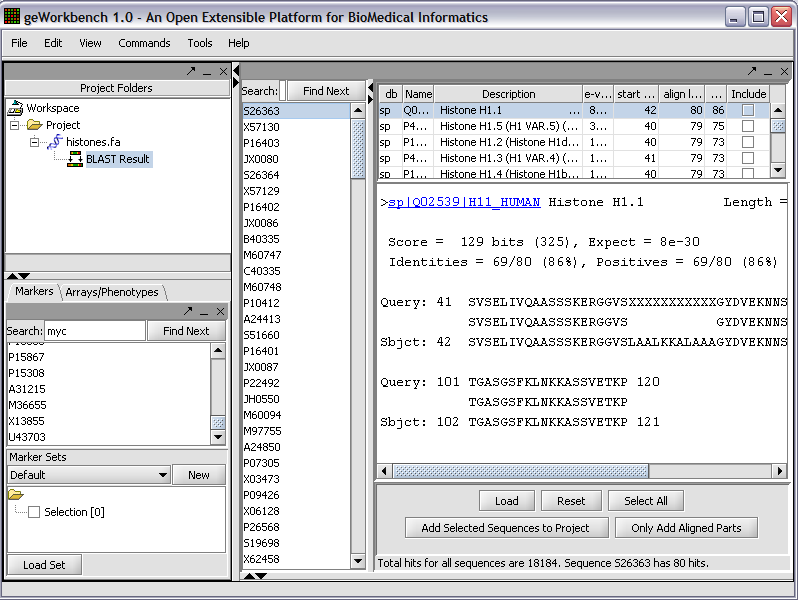
BLAST Queries
The BLAST panel runs queries against the Paracel Blast machine running at the Columbia Genome Center.