Difference between revisions of "User:Floratos/MRA"
(→Working with and viewing the analysis results) |
(→Overview) |
||
| Line 4: | Line 4: | ||
Regulatory activity in the context of specific cellular phenotypes can be modeled using interaction networks. These are graphs where nodes represent genes and an edge between two nodes A and B means that genes ''A'' and ''B'' are participants in the same regulatory activity. E.g., ''A'' can be a transcription factor for ''B''; or, ''A'' can be an miRNA that silences ''B''. Analysis of such regulatory networks [refs] has convincingly demonstrated their scale free nature which is dominated by a relatively small number of nodes with a large degree of connectivity. The genes corresponding to those nodes are known as "master regulators" and collectively orchestrate the regulatory program of the underlying cellular phenotype(s). | Regulatory activity in the context of specific cellular phenotypes can be modeled using interaction networks. These are graphs where nodes represent genes and an edge between two nodes A and B means that genes ''A'' and ''B'' are participants in the same regulatory activity. E.g., ''A'' can be a transcription factor for ''B''; or, ''A'' can be an miRNA that silences ''B''. Analysis of such regulatory networks [refs] has convincingly demonstrated their scale free nature which is dominated by a relatively small number of nodes with a large degree of connectivity. The genes corresponding to those nodes are known as "master regulators" and collectively orchestrate the regulatory program of the underlying cellular phenotype(s). | ||
| − | The master regulator analysis (MRA) component in geWorkbench combines regulatory information from interaction networks with differential expression analysis. The objective is to place differentially expressed genes within a regulatory context and identify the master regulators responsible for coordinating their regulation, thus highlighting the regulatory apparatus driving phenotypic differentiation. Specifically, given an interaction network ''I'', a master regulator gene ''A'', and two sets of microarrays representing two distinct phenotypes, MRA computes the intersection between two sets of genes: | + | The master regulator analysis (MRA) component in geWorkbench combines regulatory information from interaction networks with differential expression analysis. The objective is to place differentially expressed genes within a regulatory context and identify the master regulators responsible for coordinating their regulation, thus highlighting the regulatory apparatus driving phenotypic differentiation. Specifically, given an interaction network ''I'', a (presumed) master regulator gene ''A'', and two sets of microarrays representing two distinct phenotypes, MRA computes the intersection between two sets of genes: |
# The neighbors of ''A'' in the interaction network ''I'' (this gene set is called the '''''regulon''''' of ''A''). | # The neighbors of ''A'' in the interaction network ''I'' (this gene set is called the '''''regulon''''' of ''A''). | ||
# The set of differentially expressed genes in the array data from the two phenotypes of interest. | # The set of differentially expressed genes in the array data from the two phenotypes of interest. | ||
| − | Fisher’s exact test is then used to quantify how likely it is to encounter an intersection of the observed size by chance alone. A small p-value is taken to imply that gene ''A'' may play a significant role in | + | Fisher’s exact test is then used to quantify how likely it is to encounter an intersection of the observed size by chance alone. A small p-value is taken to imply that gene ''A'' may play a significant role in mediating the regulatory program that leads to the differential phenotypes. |
=Setting up an MRA run= | =Setting up an MRA run= | ||
Revision as of 12:58, 22 October 2009
|
Aris's Home | Enhancements | Release v1.1 | Bison Issues | MRA | Release Testing | Home Page |
Contents
Overview
Regulatory activity in the context of specific cellular phenotypes can be modeled using interaction networks. These are graphs where nodes represent genes and an edge between two nodes A and B means that genes A and B are participants in the same regulatory activity. E.g., A can be a transcription factor for B; or, A can be an miRNA that silences B. Analysis of such regulatory networks [refs] has convincingly demonstrated their scale free nature which is dominated by a relatively small number of nodes with a large degree of connectivity. The genes corresponding to those nodes are known as "master regulators" and collectively orchestrate the regulatory program of the underlying cellular phenotype(s).
The master regulator analysis (MRA) component in geWorkbench combines regulatory information from interaction networks with differential expression analysis. The objective is to place differentially expressed genes within a regulatory context and identify the master regulators responsible for coordinating their regulation, thus highlighting the regulatory apparatus driving phenotypic differentiation. Specifically, given an interaction network I, a (presumed) master regulator gene A, and two sets of microarrays representing two distinct phenotypes, MRA computes the intersection between two sets of genes:
- The neighbors of A in the interaction network I (this gene set is called the regulon of A).
- The set of differentially expressed genes in the array data from the two phenotypes of interest.
Fisher’s exact test is then used to quantify how likely it is to encounter an intersection of the observed size by chance alone. A small p-value is taken to imply that gene A may play a significant role in mediating the regulatory program that leads to the differential phenotypes.
Setting up an MRA run
Prerequisites
- First confirm that the MRA component is available in geWorkbench. If not, it can be loaded using the Component Configuration Manager.
- The MRA will be listed along with the other analysis routines within the geWorkbench Analysis pane.
- Data from an expression experiment (comprising measurements from multiple arrays/samples) has been loaded and the corresponding node has been selected withing the Project Folders:
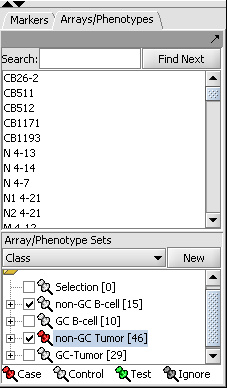
- Two array sets (each containing a distinct subset of arrays from the expression experiment) have been selected in the "Arrays/Phenotypes" component, representing the two phenotypes under investigation. One of the array sets (identified by the red-color pin) has been classified as containing the "Case" arrays while the other one contains the "Controls":
NOTE:The "Case"/"Control" assignment is needed because, in the general case, it is possible to select more than one array sets for this analysis. In that case it is necessary to explicitly designate which belong to each of the 2 phenotypes.
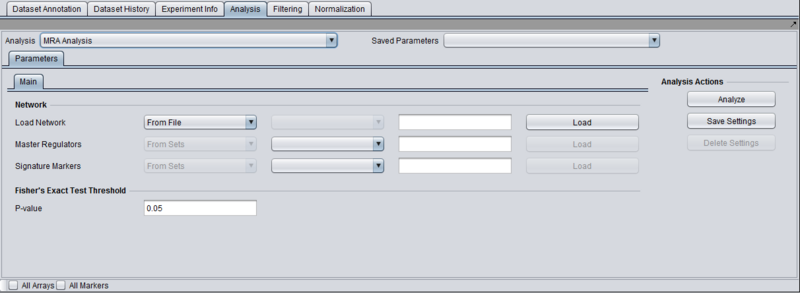
Parameters and Settings
Load Network
There are 2 ways to designate the interaction network that will be used for computing the neighbors of the candidate master regulator genes:
- From File: by choosing a file that describes a network (see example and format description below).
- From Project: by selecting a node from the project folders which represents an interaction network. Several analytical components in geWorkbench (e.g., ARACNE) produce results nodes that can be utilized for this purpose.
Master Regulators
There are 2 ways to designate the candidate master regulator genes to use:
- From File: by choosing a file that contains a list of (comma separated) marker names.
- From Sets: by selecting one among the marker sets within the “Markers” component.
T-test p-value (alpha)
Differential expression between the 2 phenotypes of interest is assessed using a t-test. The p-value provided by the user indicates the significance threshold below which a gene’s average expression is presumed to be significantly different in the 2 sets of arrays (cases and controls). Additional parameter settings affecting the execution of the t-test are defined within the “T-test” subtab. The parameters specified there are exactly the same as those used for the differential expression t-test analysis (see description in the corresponding tutorial.
Working with and viewing the analysis results
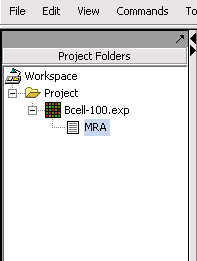
Following the successful completion of the MRA computation, a result node appears in the Project Folder area of the geWorkbench interface, under the microarray experiment node used for the computation:
The results of the analysis can be visualized in the MRA Viewer component by selecting the result node.
MRA Results Viewer
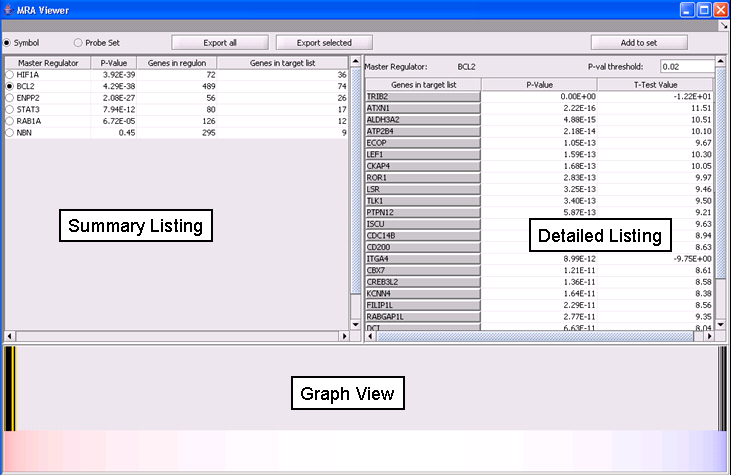
The MRA viewer is structured in 3 distinct areas.
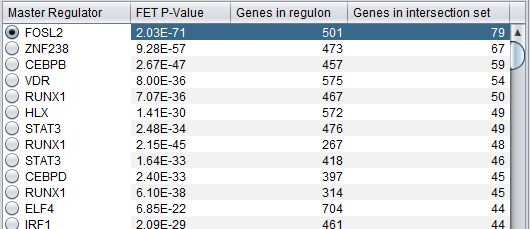
Summary Listing
This is a table with one row per each candidate master regulator specified in the parameters panel.
Each row contains 4 columns:
- Master Regulator: This is either the master regulator gene name or the marker/probeset name identifying the corresponding array feature (depending on the selection of 2 radio buttons titled “Symbol” and “Probe set”).
- P-value: the p-value of fisher’s exact test. See below for detailed description of this computation.
- Genes in regulon: The number of genes in the regulon of the master regulator (this comprises the set of genes that are first neighbors of the master regulator in the interaction network specified in the parameters panel).
- Genes in target list: The number of differentially expressed genes that are also members of the regulon.
The contents of the table can be ordered across any column, by clicking on the column name.
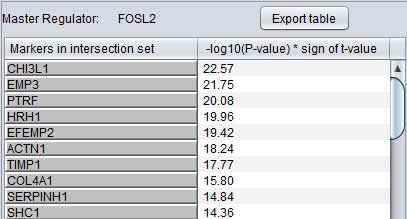
Detailed Listing
By clicking on the radio button associated with a master regulator in the Summary table, it is possible to display the complete listing of the differentially expressed genes intersecting the regulon of that master regulator.
The genes are displayed in a table with the following columns:
- Genes in target list: contains the names of the genes in the intersection set. Either the gene name or the marker/probe set name is used.
- P-value: the p-value of the t-test statistic for this marker, computed over the 2 sets of arrays (cases versus controls).
- T-test value: The actual value of the t-test statistic for the gene. A positive value indicates that the mean expression of the gene is higher in cases that in controls (a negative value has the opposite meaning).
The “P-val Threshold” parameter (located above the table) can be used to limit the number of target genes displayed. Specifically, only genes with a p-value less than the specified cutoff will be shown (if “P-val Threshold” is left empty, then all target genes from the intersection set of the currently selected TF are displayed). Additionally functionality is made available through the following buttons:
- Add to Set: creates a set containing the markers that correspond to the genes currently displayed in the table. The new marker set appears in the Markers component and is named after the master regulator.
- Export selected: same as “Add to Set” but instead of being added to a marker set, the markers are stored into a file.
- Export all: stores into a file information for each of the master regulators (instead of only the one currently selected in the “Summary” view). Specifically, for each master regulator, the file lists all the markers that belong both in the list of differential expressed genes as well as in the master regulator’s regulon.
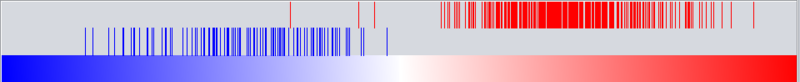
Graph View
For a given master regulator A and the intersection between its regulon and the set of differentially expressed genes, the graph view helps assesing if the intersection genes are preferentially over-expressed in the cases versus the controls. The biological motivation comes from observing [ref] that regulators with multiple targets tend to affect the expression level of (most of) their targets in one particular direction: they either promote their expression or inhibit it; but they rarely do both equally.
The red-blue gradient at the bottom of the graph represents the range between the highest (red) and the lowest (blue) t-test statistic recorded among the differentially expressed genes. The vertical bars correspond to the genes displayed in the table under the “Detailed listing” portion of the interface, i.e., the intersection between the differentially expressed genes and the master regulator A currently selected within the “Summary listing” table. The relative location of a bar on the gradient represents the t-test statistic recorded for the corresponding gene. Further, the color of each bar provides information about the correlation between the expression levels of the target gene and the master regulator A (correlations are computed using Pearson’s r, using data from all microarrays in the experiment): black means that the two genes are positively correlated (r > 0) while orange means that correlation is negative (r < 0).
Dataset History
Each results node stores the parameter settings used to setup the corresponding MRA run. The specific parameter values can be inspected within the Dataset History component, after clicking on the MRA results node in the Project Folders pane.