Screenshots
Contents
geWorkbench Configuration
=Component Configuration Manager
Individual components can be loaded as needed.
Microarray Data Displays
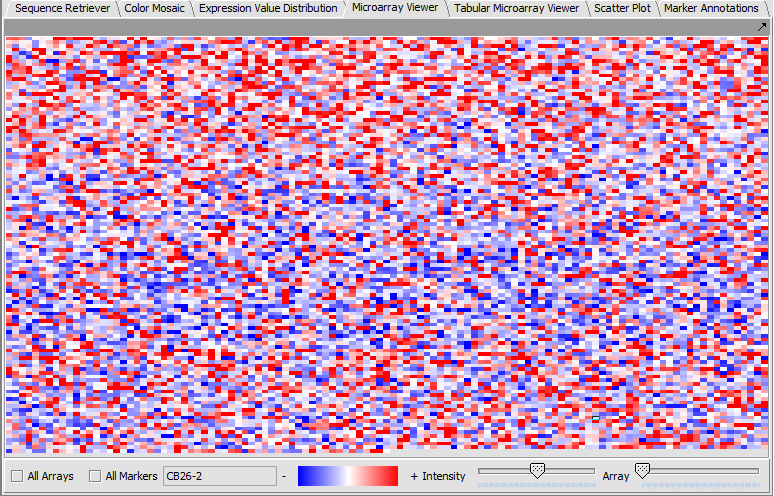
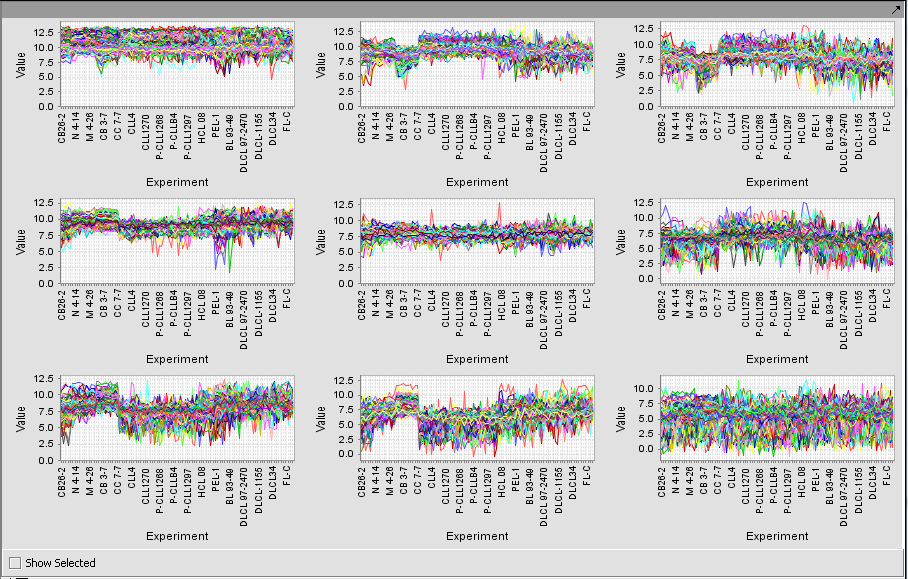
Microarray Viewer
The Microarray Viewer displaying marker values for selected array.
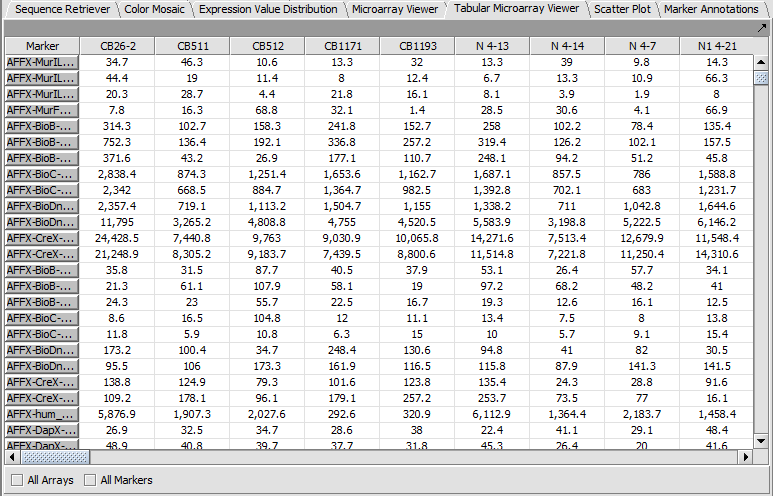
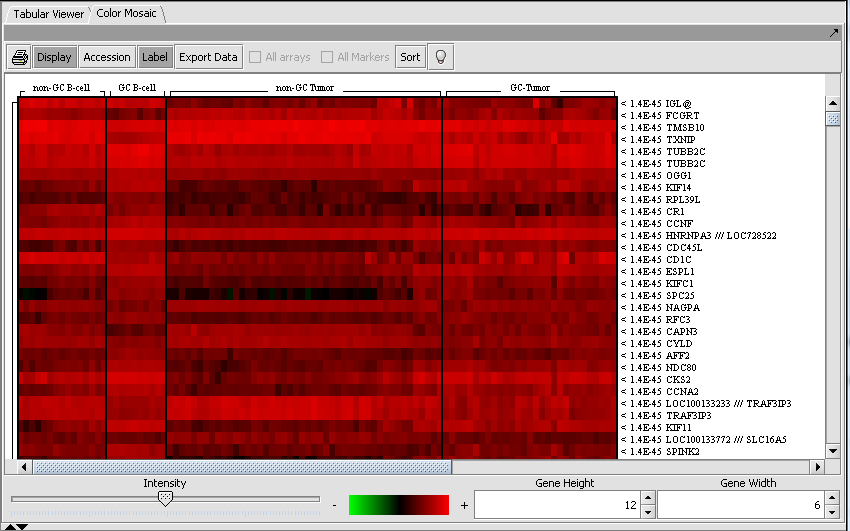
Tabular Microarray Viewer
The Tabular Microarray Viewer displays expression values in spreadsheet format.
CEL Image Viewer
Allows viewing of Affyemtrix CEL files.
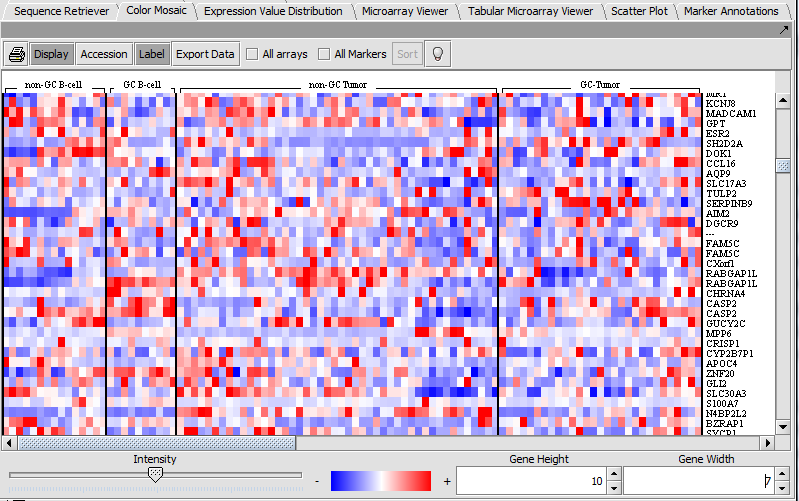
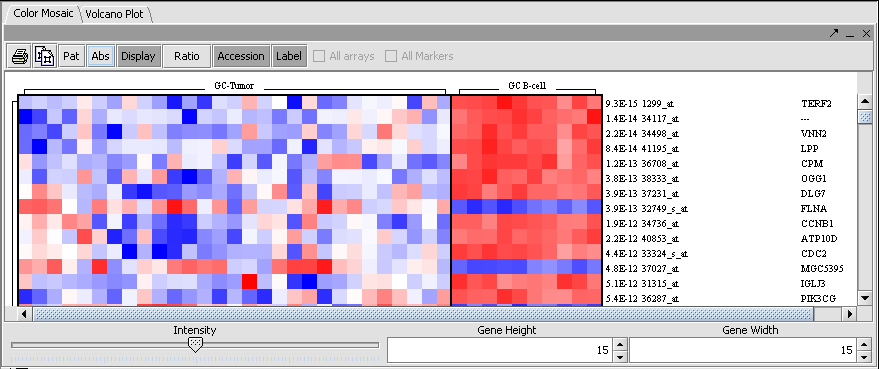
Color Mosaic
The Color Mosaic component displaying selected arrays, group designation and marker names.
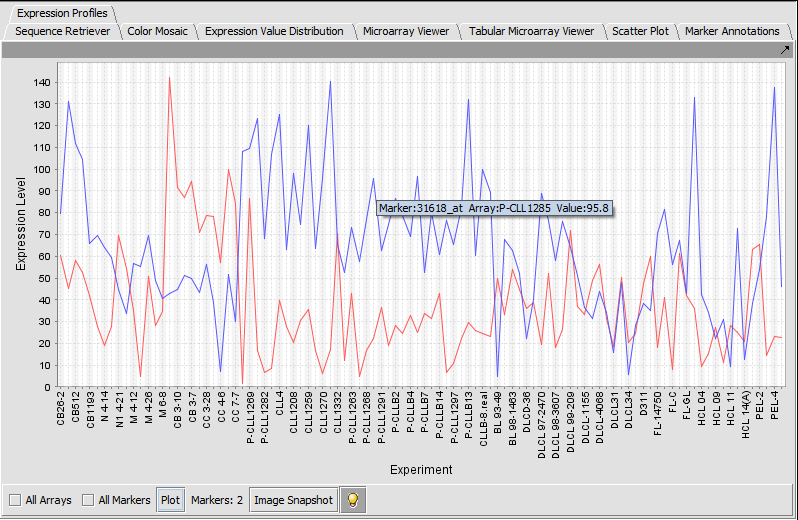
Expression Profile
Expression Profile plotting values for selected markers and arrays.
TP53 (31618) and PTTG2 (31631_f_at)
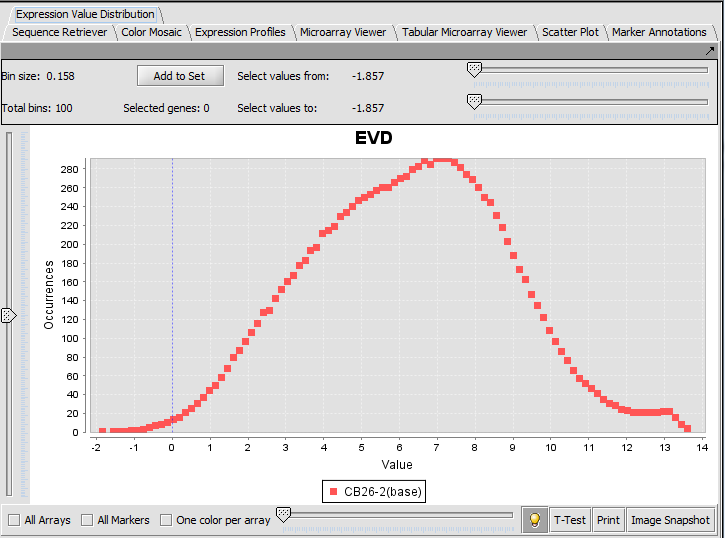
Expression Value Distribution
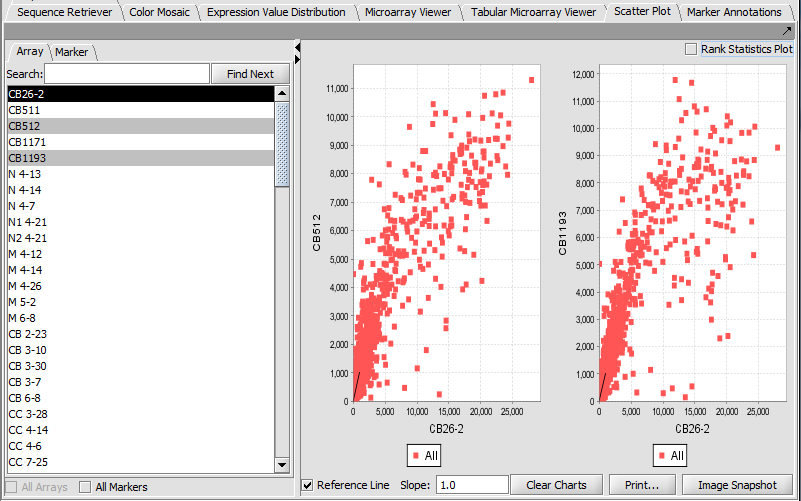
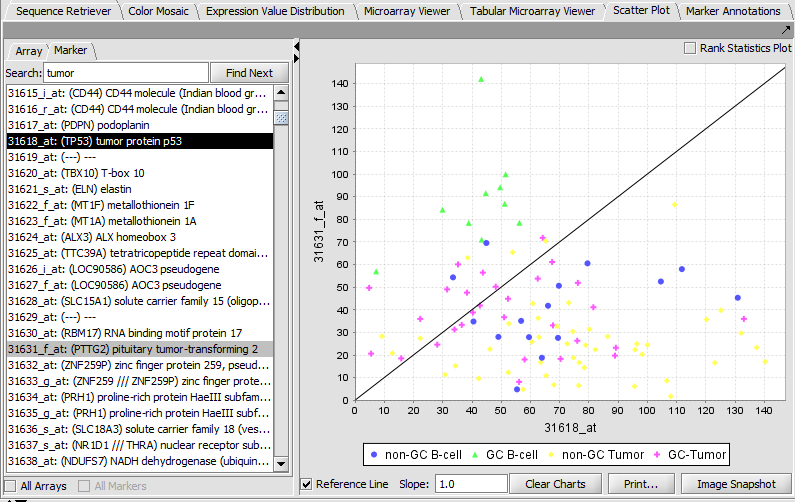
Scatter Plot
Compare multiple markers or arrays with the standard Scatter Plot analysis.
Array vs Array
Marker vs Marker
Gene and Pathway Annotations
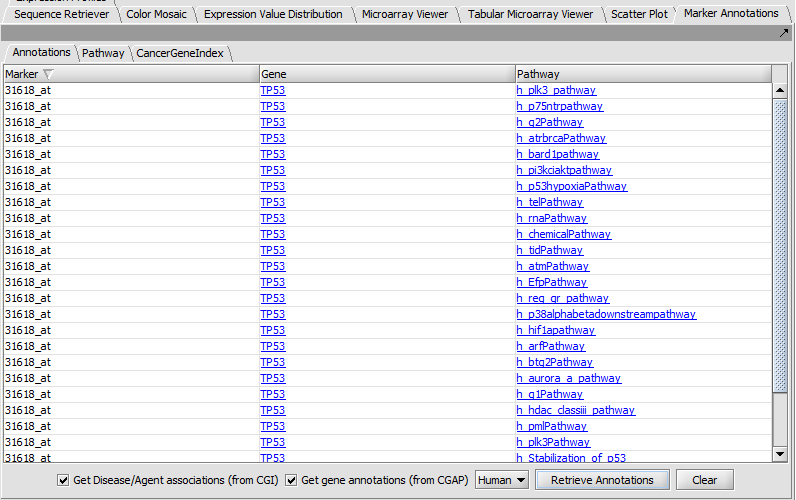
Marker Annotations
Retrieve and display gene and pathway information from CGAP and Cancer Gene Index (CGI).
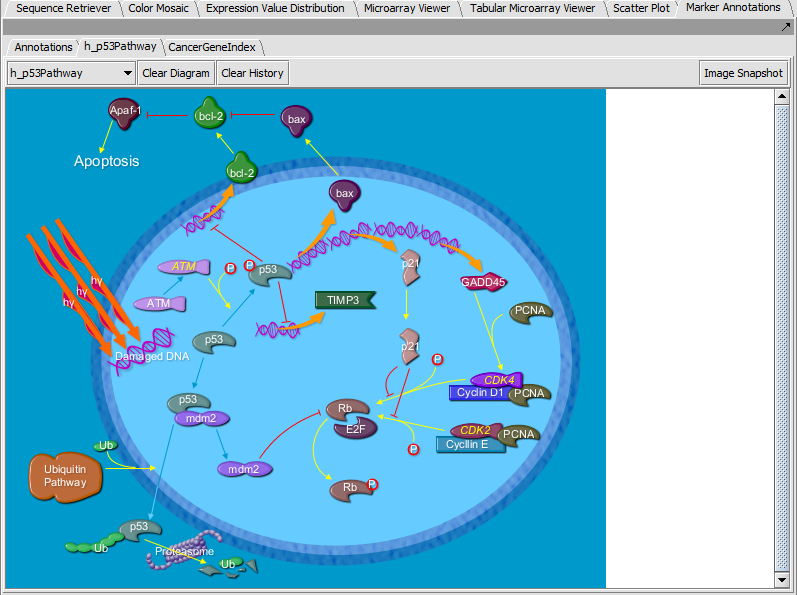
Marker Annotations - BioCarta Pathways
Displays BioCarta images retrieved from caBIO.
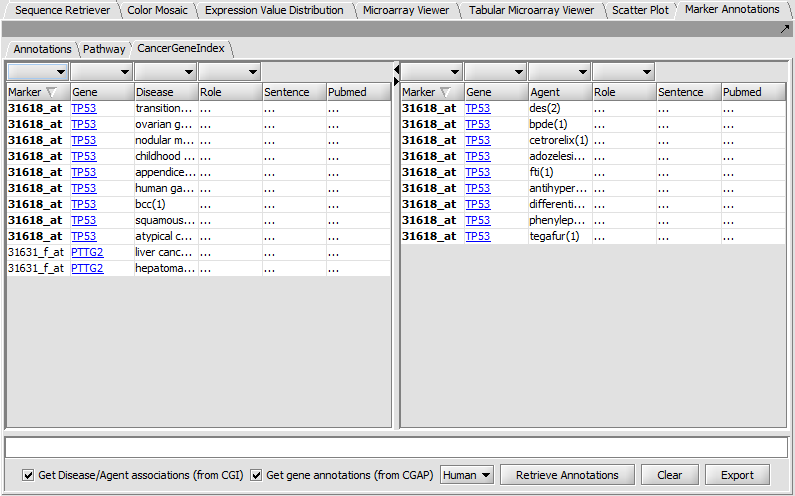
Marker Annotations - Cancer Gene Index
Displays literature citations from the Cancer Gene Index project.
Statistical tests and clustering
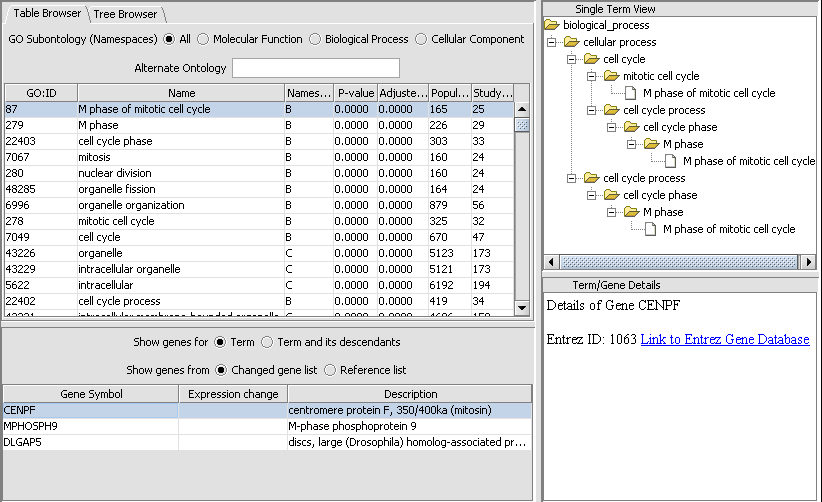
Gene Ontology Term Over-representation Analysis
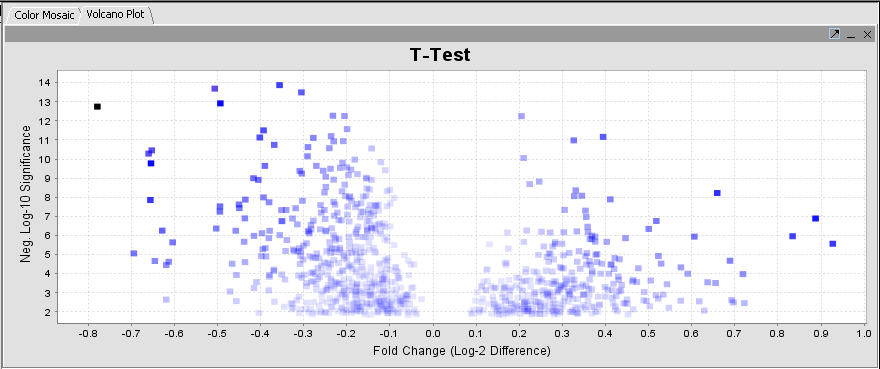
T Test
Volcano Plot
A t-test result display on a "volcano plot": Log significance vs log fold change.
Color Mosaic
The t-test result can also be displayed as a color mosaic.
(Visualization preference setting: Relative)
Analysis of Variance (ANOVA)
Detects markers for which a statistically significant difference exists in a data set containing multiple classes of samples.
(Visualization preference setting: Absolute)
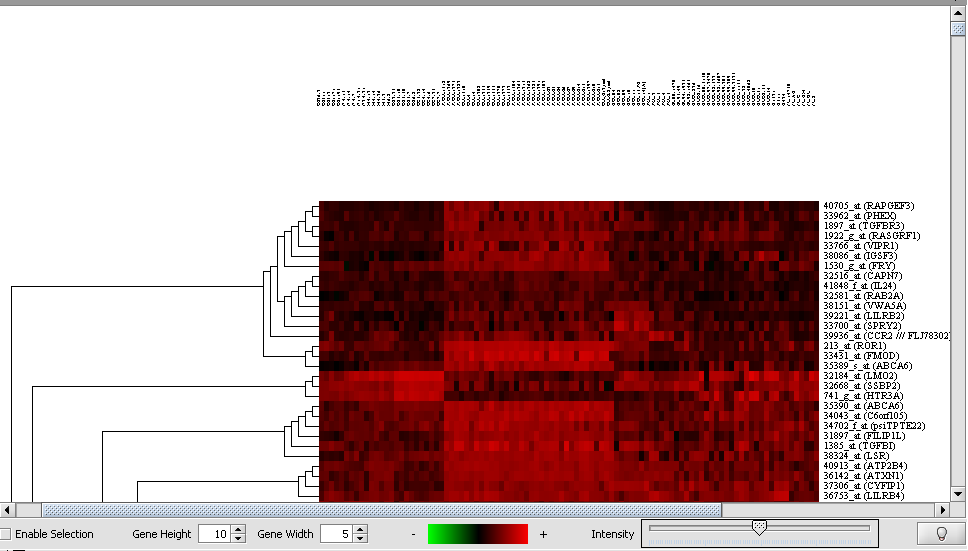
Hierarchical Clustering Dendrogram
A Dendrogram displays the results of the Hierarchical clustering analysis.
SOM Clustering
Self Ordered Map clustering results are displayed as series of expression profiles corresponding to discovered groupings.
Sequence Analysis / Pattern Discovery
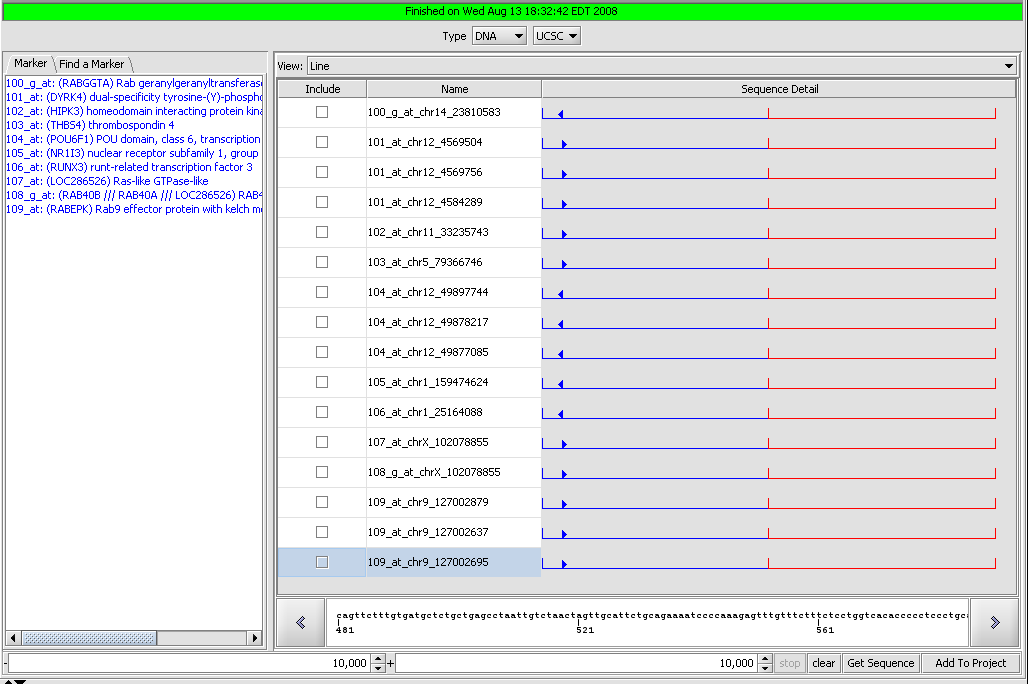
Sequence Retriever
Retrieve genomic and protein sequences for selected markers. Retrieved sequences can be individually selected and added to the project as new data nodes.
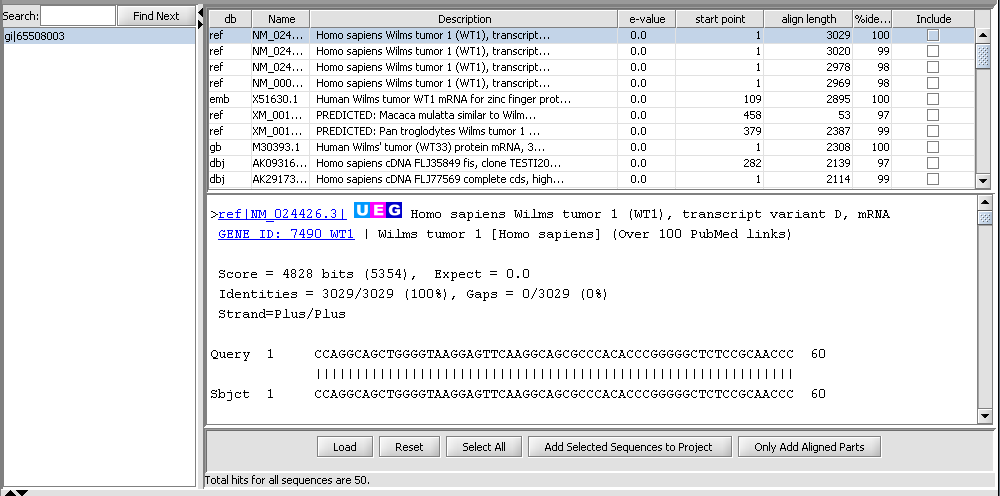
BLAST Queries
The Sequence Alignment component submits BLAST jobs to the NCBI server and displays the results such that individual hits can be used in further analysis steps.
Pattern Discovery
Use the SPLASH algorithm to discover sparse amino or nucleic acid patterns in a loaded sequence.
Motif discovery and display
The Pattern Discovery component itself with results displayed in the sequence viewer.
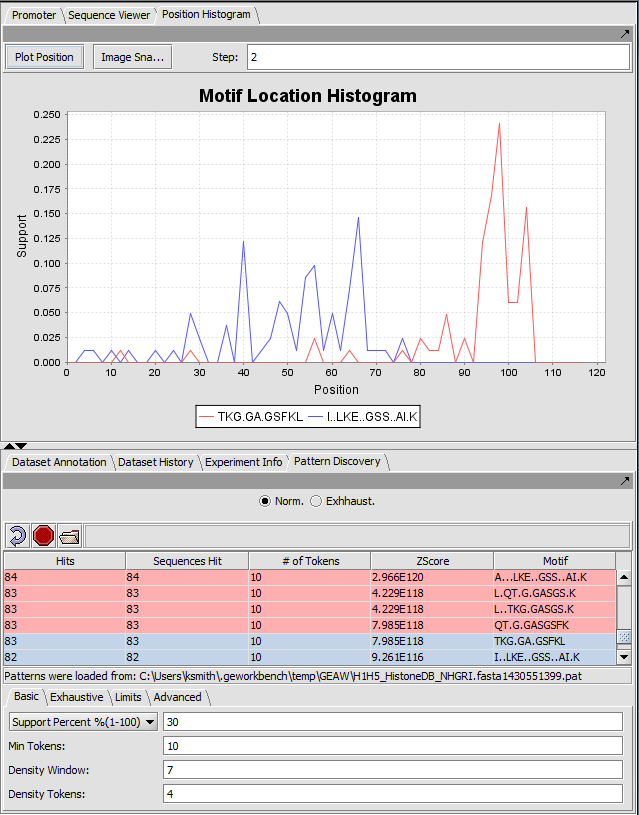
Position Histogram
The Pattern Discovery component with results displayed as histogram of support for selected discovered motifs across the sequence data set. Support indicates what fraction of the seqeunces are matched by the motif at a within a sliding window about a given location.
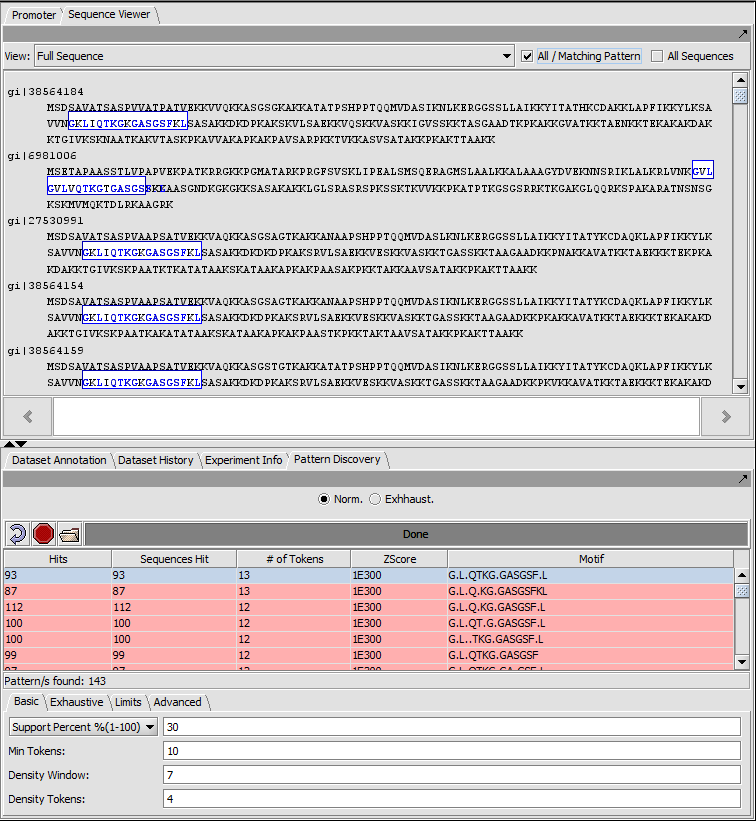
Promoter
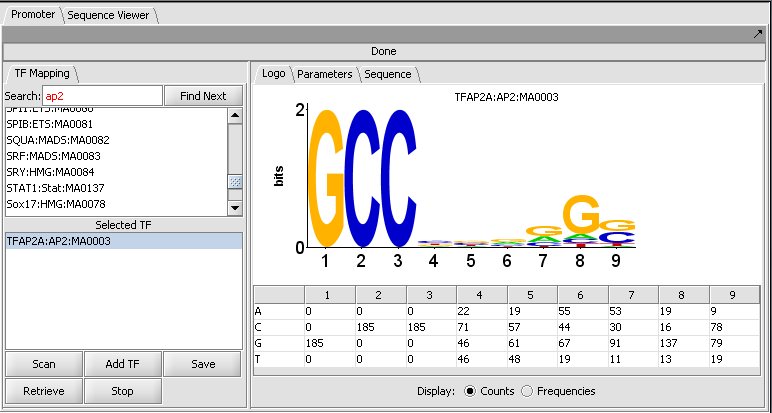
Individual motifs from the JASPAR Transcription Factor Binding Profile Database can be scanned against loaded genomic sequences.
Motif selection and Logo display
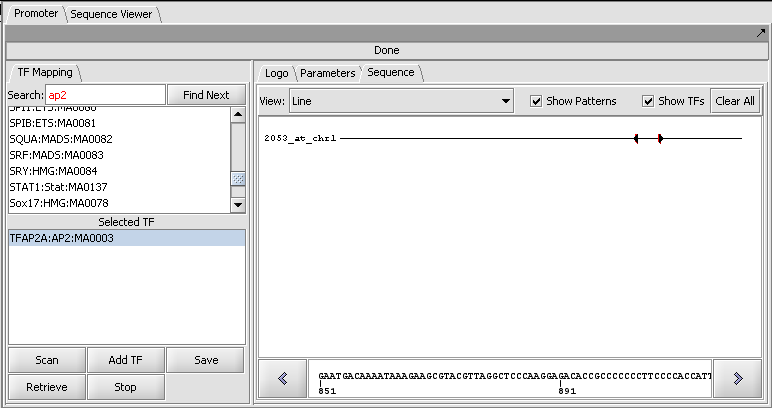
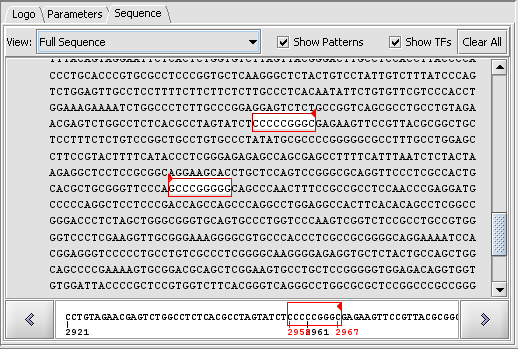
Result of a scan against a single sequence
Sequence-level display of match
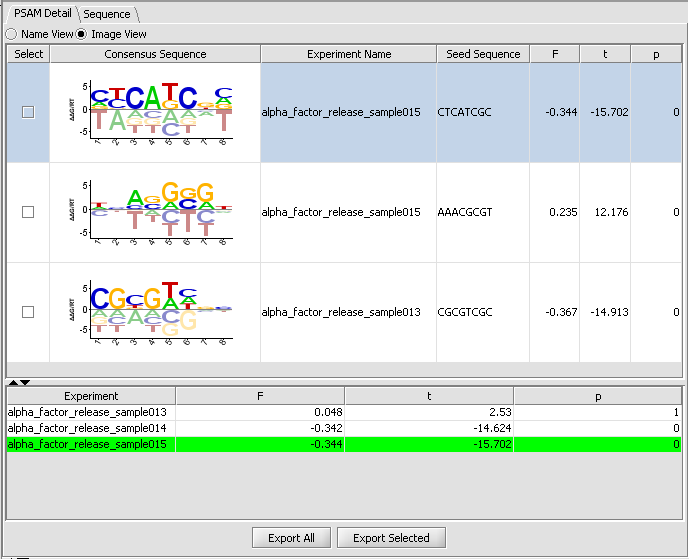
MatrixREDUCE
MatrixREDUCE is a tool for inferring the binding specificity and nuclear concentration of transcription factors from microarray data.
Network Discovery and Visualization
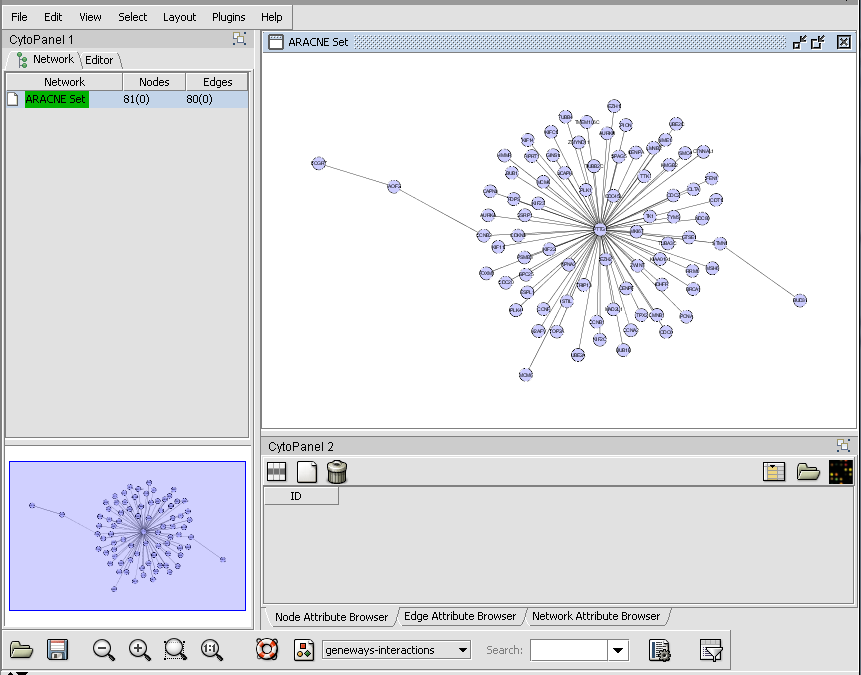
Cytoscape - ARACNe Network display
The adjacency matrix generated by an ARACNe network reverse engineering run displayed in Cytoscape.
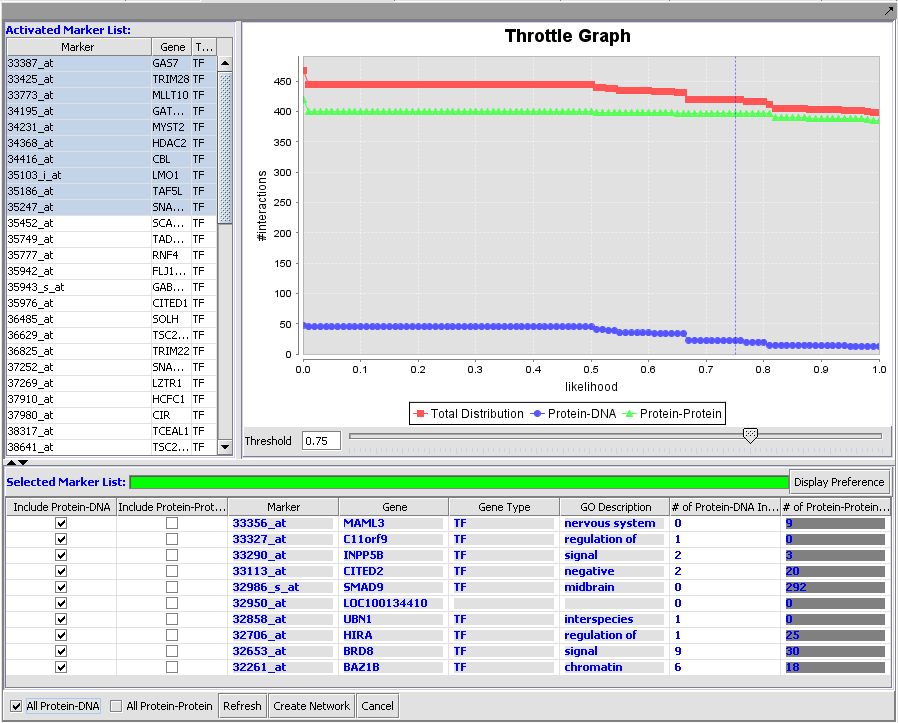
Cellular Network Knowledge Base
Results of queries against the CNKB can be filtered based on confidence values using the throttle graph.
Query results and Throttle Graph
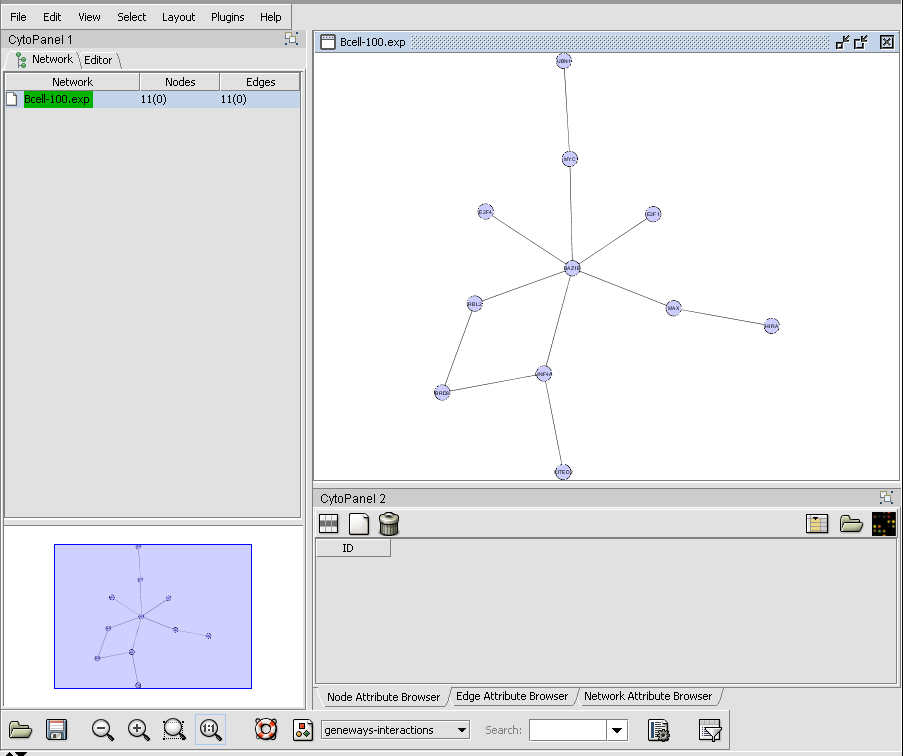
CNKB query results displayed in Cytoscape
Display of protein-DNA interactions produced with throttle graph set at 75% confidence in above picture.