FAQ
Contents
- 1 Using geWorkbench
- 1.1 Q. Can I run geWorkbench on a 64-bit Windows system?
- 1.2 Q. I get a Java error when I try to start geWorkbench.
- 1.3 Q. Where can I obtain the latest annotation files for my microarray platform?
- 1.4 Q. How do I increase the amount of memory available to Java to run geWorkbench?
- 1.5 Q. Is there a shortcuts menu for available commands?
- 1.6 Q. geWorkbench appears to be frozen.
- 1.7 Q. How can I download the geWorkbench source?
- 1.8 Q. How do I turn on logging in installer-based versions of geWorkbench?
- 1.9 Q. How can I reference geWorkbench in a publication?
- 1.10 Q. What differences are there in how genes without symbols are handled in GO Terms analysis vs a CNKB query?
Using geWorkbench
Q. Can I run geWorkbench on a 64-bit Windows system?
A. Yes. As of release 2.0.0, geWorkbench has been tested on both 64-bit Windows 7/Vista operating systems, as well as 32-bit Windows XP. The formal release testing for Windows is done using the 32-bit Java JRE6. However, geWorkbench has also been tested with the 64-bit Java JRE6 on the 64-bit Windows 7 platforms, and only minor compatibility issues have been found. These are listed in the Known Issues section.
The prebuilt installers with JRE6 all include the 32-bit JRE. If you wish to try the 64-bit JRE, please use a version of the geWorkbench installer which does not include the JRE. Further instructions can be found on the Download and Installation page.
Q. I get a Java error when I try to start geWorkbench.
A. This is almost always caused by the Sun Java JRE not being installed or not being found (when you use a version of geWorkbench that does not include the JRE). You can either try a geWorkbench installer that includes the JRE, or make sure that an appropriate JRE is installed on your system.
Q. Where can I obtain the latest annotation files for my microarray platform?
A. Affymetrix annotation files can be downloaded from their support site, at www.affymetrix.com. A free Netaffx account sign-up is required.
There are several ways to reach the annotation files, two of which are shown next:
1. The following link will take you directly to a list of most (but not all) current annotation files.
http://www.affymetrix.com/support/technical/annotationfilesmain.affx
2. Some arrays, for example the HG-U95Av2 array used in some geWorkbench tutorials (for the BCell-100.exp data set), are not shown in the above list. However, the files can be obtained from the catalog of individual array products at the following link:
http://www.affymetrix.com/support/technical/byproduct.affx?cat=arrays
For the HG-U95Av2 annotation file, browse down to 3' Gene Expression Analysis and look for Human Genome Arrays. Then find the section "Current NetAffx Annotation Files".
The file format required is CSV (comma separated values).
Q. How do I increase the amount of memory available to Java to run geWorkbench?
A. It depends on how you are running geWorkbench.
1. If you are running a packaged distribution of geWorkbench (created using InstallAnywhere), there is a file in the geWorkbench root directory called UILauncher.lax. There is a line there which specifies the Java heap size:
lax.nl.java.option.java.heap.size.max=1073741824
Here it is shown set to 1 GB. You can experiment with increasing this, subject to the amount of memory in your machine and demands on it from other applications.
2. If you are running geWorkbench from the Generic or source distributions using Ant, you can edit the build.xml file found in the geWorkbench root directory to alter the memory requested using the variable jvmarg:
<target name="run" depends="init" description="Runs geWorkbench.">
<java fork="true" classname="org.geworkbench.engine.config.UILauncher">
<jvmarg value="-Xmx1024M"/>
<jvmarg value="-Djava.library.path=lib"/>
<arg value="all_release.xml"/>
<classpath refid="run.classpath"/>
</java>
</target>
Here it is shown requesting 1 GB.
A. Yes! - on the PC, you can use F12.
Q. geWorkbench appears to be frozen.
A. Sometimes, a "modal" dialog box can appear but by chance get hidden behind other windows (if the user clicks on some other window without noticing the dialog box). The hidden dialog box is waiting for user interaction, and geWorkbench is waiting for the dialog to be dismissed, so the application is unresponsive. This is a feature of how Java works. One way to see if this has happened is to minimize all open windows on your desktop, for all applications, and the maximize the geWorkbench window. If there is an open dialog box, it should now appear in front of the geWorkbench main window. (c.f. Mantis entry 1959).
Q. How can I download the geWorkbench source?
A. NOTICE - to avoid a huge download, make sure to follow all of the below instructions to when checking out the development source code.
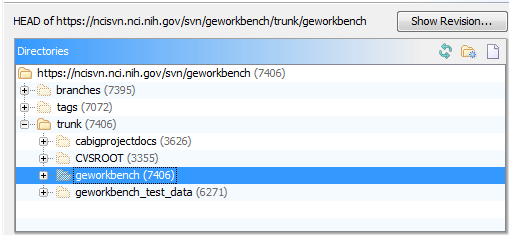
- The full URL for the development trunk of geworkbench at the NCI SVN site is https://ncisvn.nci.nih.gov/svn/geworkbench/trunk/geworkbench.
- If you are using a graphical SVN client, such as SmartSVN,
- the primary URL is https://ncisvn.nci.nih.gov/svn/geworkbench/,
- and you should navigate to trunk/geworkbench. This process is illustrated below using SmartSVN:
- For just checking out the code, you can use the username and password of anonymous/anonymous.
- To commit changes, you must be a registered member of the geWorkbench development project and logged in using an NCI LDAP account.
Whichever method is used, this is a source distribution which you will need to compile yourself:
- you must have the Java JDK 6 (or higher) installed on your machine - see http://java.sun.com, under the "Java SE" link.
- you must make sure that the value of the environment variable JAVA_HOME is the directory where the JDK is installed.
Note that Sun recommends adding the Java JDK bin directory to your machine's PATH variable. Under Windows, "Typically this full path looks something like C:\Program Files\Java\jdk1.6.0_<version>\bin." For example, the current release installs in "C:\Program Files\Java\jdk1.6.0_20". Full instructions can be found on the Java site.
If you have Ant installed on your machine, you can just type "ant run" in the geworkbench root directory and the program will be built and run. Ant can be downloaded from http://ant.apache.org/. Note that installing Ant involves manually adding the Ant bin directory to the PATH variable, setting the ANT_HOME directory, and optionally setting the JAVA_HOME directory.
If you do not have Ant, you can just execute one of the provided launch scripts, also found the in geworkbench root directory. They are:
- Windows: launch_geworkbench.bat
- Linux/Unix/Mac: launch_geworkbench.sh
Q. How do I turn on logging in installer-based versions of geWorkbench?
Copies of geWorkbench installed using the InstallAnywhere-generated install file contain a configuration file in their root directory called UILauncher.lax. Make the changes like the following to turn on logging to files:
# LAX.STDERR.REDIRECT # ------------------- # leave blank for no output, "console" to send to a console window, # and any path to a file to save to the file
lax.stderr.redirect=log/stderr.log
# LAX.STDOUT.REDIRECT # ------------------- # leave blank for no output, "console" to send to a console window, # and any path to a file to save to the file
lax.stdout.redirect=log/stdout.log
Q. How can I reference geWorkbench in a publication?
A1: geWorkbench is described in the following publication:
http://www.ncbi.nlm.nih.gov/pubmed/20511363
Floratos A, Smith K, Ji Z, Watkinson J, Califano A. (2010). geWorkbench: an open source platform for integrative genomics. Bioinformatics 26(14):1779-80. Epub 2010 May 28.
A2: Further support statement:
- "Analysis was carried out using geWorkbench (http://www.geworkbench.org), a free open source genomic analysis platform developed at Columbia University with funding from the NIH Roadmap Initiative (1U54CA121852-01A1) and the National Cancer Institute".
Q. What differences are there in how genes without symbols are handled in GO Terms analysis vs a CNKB query?
A. In the GO Terms analysis component, only genes with a gene symbol are used in the analysis, and only genes with a gene symbol will appear in the list of genes associated with a selected term. By contrast, in the CNKB component, markers which match the particular id (e.g. Entrez ID) used in the query are returned even if they do not have an associated gene symbol. (Mantis issue #2478).