Consensus Clustering
Overview
This component allows geWorkbench to run the Consensus Clustering on a GenePattern server. The user must supply the URL of an available GenePattern server. All results are returned to geWorkbench, and the clustered arrays or markers are available as sets for further analysis.
For information on the Consensus Clustering algorithm, please see the GenePattern documentation at GenePattern Analysis:Modules.
Parameters
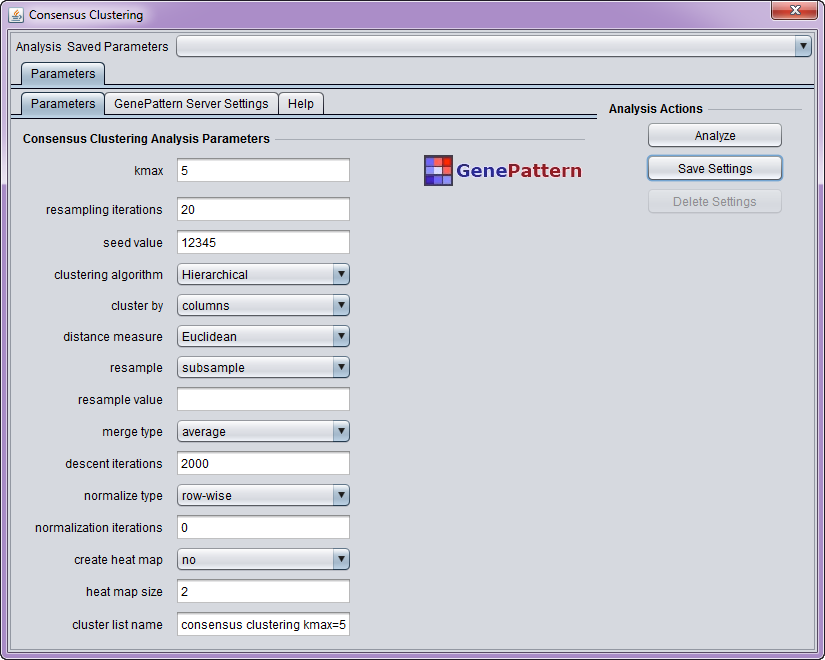
- kmax - Try K=2,3,...,kmax clusters (must be > 1)
- resampling iterations - Number of resampling iterations
- seed value - Random number generator seed
- clustering algorithm - Type of clustering algorithm. Choices are
- Hierarchical
- SOM
- NMF
- KMeans
- cluster by - Whether to cluster by rows/genes or columns/experiments
- distance measure - Distance measure. Either
- Euclidean
- Pearsons
- resample - resampling scheme (one of 'subsample[ratio]', 'features[nfeat]', 'nosampling')
- resample value - ratio or nfeat can be optionally input as indicated above.
- merge type - Ignored when algorithm other than hierarchical selected
- Average
- Complete
- Single
- descent iterations - Number of SOM/NMF iterations
- output stub - Stub prepended to all the output file names
- normalize type - row-wise, column-wise, both, none.
- normalization iterations - number of row/column normalization iterations (supersedes 'normalize type')
- create heat map - Whether to create heatmaps (one for each cluster number): no or yes
- heat map size - point size of a consensus matrix's heat map (between 1 and 20)
- cluster list name - name for new geWorkbench marker/array context in which to store clusters.
References
References: • S. Monti, et al. “Consensus Clustering: A resampling-based method for class discovery and visualization of gene expression microarray data”, Machine Learning Journal, 52(1-2):91-118, 2003.