Difference between revisions of "Cupid"
(→Inputs) |
(→Parameters) |
||
| Line 16: | Line 16: | ||
===Parameters=== | ===Parameters=== | ||
| − | The CUPID | + | * '''Server URL''' - The URL of the CUPID service (at Columbia). The user should not need to change this. |
| − | Type | + | * '''Query Type''' - CUPID results can be queried either by RefSeq ID or miRNA id. |
| − | Value | + | ** RefSeq ID - query by RefSeq ID. |
| + | ** miRNA ID - query by miRNA ID. | ||
| + | * '''Query Value''' - The RefSeq or miRNA ID on which to query the database. | ||
===Results=== | ===Results=== | ||
Revision as of 12:43, 13 January 2014
Overview
CUPID generates information that can help predict if a gene is a target of a specific miRNA. The Cupid service provides a simple query interface to a database of precalculated Cupid results. The results can be searched either by miRNA ID or by RefSeq gene ID.
For each miRNA M the algorithm uses TargetScan [11], PITA [12], and MIRANDA [13], at their default settings, to predict target sites of M in 3' UTRs of REFseq (hg19) transcripts. Each predicted site seed is scored according to its conservation across 46 vertebrate genomes (http://hgdownload.cse.ucsc.edu/goldenPath/hg19/phastCons46way). Site distances from 5' and 3' ends of the 3'UTR are also annotated. Scores are normalized and input into a support vector machine tool (LIBSVM, http://www.csie.ntu.edu.tw/~cjlin/libsvm/faq.html#f203) to train against 684 validated miRNA targets (the “gold standard”), procured from http://mirecords.biolead.org and TRANSFAC. Site seeds classified by LIBSVM together with the gold standard interaction set are considered positive, otherwise they are considered negative. The output produced by ideal includes:
- A list of sites where an miRNA is predicted to match the 3’ UTR region of a REFseq gene. For each site the following information is listed:
- The related TargetScan, PITA and MIRANDA scores.
- The site’s conservation score.
- The site’s gold standard classification score.
- An overall probability of being a true target site (integrating all the above scores).
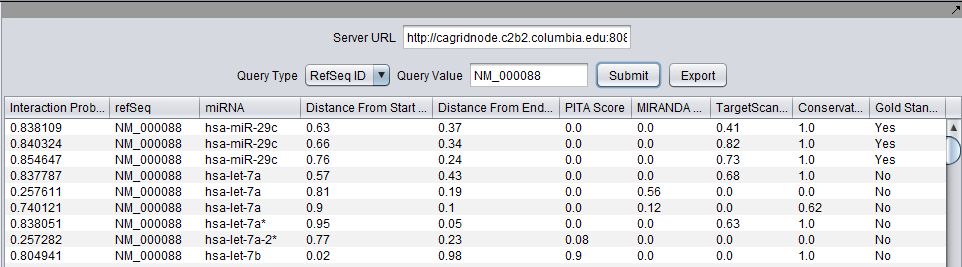
Using the CUPID Interface
Parameters
- Server URL - The URL of the CUPID service (at Columbia). The user should not need to change this.
- Query Type - CUPID results can be queried either by RefSeq ID or miRNA id.
- RefSeq ID - query by RefSeq ID.
- miRNA ID - query by miRNA ID.
- Query Value - The RefSeq or miRNA ID on which to query the database.
Results
The CUPID output lists all the sites where an miRNA is predicted to match the 3’ UTR region of a REFseq gene. Each result line represents how well a miRNA (identified by the third column of a line) is predicted to match the 3’ UTR region of a gene (identified by the second column of the line, via its refSeq id). More precisely, each column in a line captures the following info (columns below are listed in the order in which they appear in the CUPID output file):
- Interaction Probability - overall probability that the miRNA matches the 3' UTR region of the gene.
- refSeq id - refSeq idof the target gene.
- miRNA - identifier of the miRNA whose matching potential against the target gene is being assessed.
- Distance from Start of UTR - location of the starting site of the match of the miRNA sequence, on the gene’s 3’ UTR region. Distances are normalized by UTR length, e.g., 0.33 means that the beginning of the match site is 1/3 of the UTR length from its start.
- Distance from End of UTR - location of the ending site of the match of the miRNA sequence, on the gene’s 3’ UTR region. Distances are normalized by UTR length, e.g., 0.67 means that the ending of the match site is 2/3 of the UTR length from its start.
- PITA Score - PITA score for the miRNA-target site match.
- MIRANDA Score - MIRANDA score for the miRNA-target site match.
- TargetScan Score - TargetScan score for the miRNA-target site match.
- Conservation Score - Conservation score of the sequence region of the miRNA match (computed against conservation across 46 vertebrate genomes).
- Gold Standard Classification - "Yes" means that the site is classified as a gold standard; "No" that it is not).
Below are example lines from the output file produced by the CUPID code:
0.805531 NM_000034 hsa-miR-122 0.16 0.84 0.92 0.74 0.93 0.78 1 0.897525 NM_000034 hsa-miR-122 0.17 0.83 0.92 0.74 0.93 1.00 1 0.843500 NM_000038 hsa-miR-135a 0.07 0.93 0.00 0.00 0.93 1.00 1 0.836859 NM_000038 hsa-miR-135b 0.07 0.93 0.00 0.00 0.94 1.00 1 0.740299 NM_000059 hsa-miR-146a 0.65 0.35 0.00 0.29 0.90 0.00 1 0.762785 NM_000059 hsa-miR-146a 0.66 0.34 0.00 0.29 0.90 0.00 1 0.837864 NM_000076 hsa-miR-221 0.13 0.87 0.00 0.50 0.70 0.67 1 0.736315 NM_000076 hsa-miR-221 0.68 0.32 0.00 0.35 0.00 0.51 1 0.855172 NM_000076 hsa-miR-222 0.13 0.87 0.00 0.11 0.69 0.67 1 0.838109 NM_000088 hsa-miR-29c 0.63 0.37 0.00 0.00 0.41 1.00 1
However, the actual service uses pipe characters “|” rather than tabs as the delimiter:
0.838109|NM_000088|hsa-miR-29c|0.63|0.37|0.0|0.0|0.41|1.0|1| 0.840324|NM_000088|hsa-miR-29c|0.66|0.34|0.0|0.0|0.82|1.0|1| 0.854647|NM_000088|hsa-miR-29c|0.76|0.24|0.0|0.0|0.73|1.0|1| 0.837787|NM_000088|hsa-let-7a|0.57|0.43|0.0|0.0|0.68|1.0|0| 0.257611|NM_000088|hsa-let-7a|0.81|0.19|0.0|0.56|0.0|0.0|0| 0.740121|NM_000088|hsa-let-7a|0.9|0.1|0.0|0.12|0.0|0.62|0| 0.838051|NM_000088|hsa-let-7a*|0.95|0.05|0.0|0.0|0.63|1.0|0| 0.257282|NM_000088|hsa-let-7a-2*|0.77|0.23|0.08|0.0|0.0|0.0|0| 0.804941|NM_000088|hsa-let-7b|0.02|0.98|0.9|0.0|0.0|1.0|0| 0.837787|NM_000088|hsa-let-7b|0.57|0.43|0.0|0.0|0.68|1.0|0|