Home
Contents
Quick Start
Please see the Quick Start guide to geWorkbench to see how to get started using geWorkbench right away. We are continuing to develop new material for this guide.
Overview
Welcome to geWorkbench. The current version is 2.2.0, released May 24th, 2011.
Note - see Known Issues to download the new Fold Change component. It was omitted from release 2.2.0.
The latest Release Notes and downloads can be obtained from https://gforge.nci.nih.gov/frs/?group_id=78. Installation instructions can be found on the Download and Installation page of this Wiki.
geWorkbench (genomics Workbench) is a Java-based open-source platform for integrated genomics. Using a component architecture it allows individually developed plug-ins to be configured into complex bioinformatic applications. At present there are more than 70 available plug-ins supporting the visualization and analysis of gene expression and sequence data. Example use cases include:
- loading data from local or remote data sources.
- visualizing gene expression, molecular interaction networks, protein sequence and protein structure data in a variety of ways.
- providing access to client- and server-side computational analysis tools such as t-test analysis, hierarchical clustering, self organizing maps, regulatory neworks reconstruction, BLAST searches, pattern/motif discovery, etc.
- validating computational hypothesis through the integration of gene and pathway annotation information from curated sources as well as through Gene Ontology enrichment analysis.
geWorkbench is the Bioinformatics platform of MAGNet, the National Center for the Multi-scale Analysis of Genomic and Cellular Networks (one of the 7 National Centers for Biomedial Computing funded through the NIH Roadmap). Additionally, geWorkbench is supported by caBIG®, NCI's cancer Biomedical Informatics Grid initiative.
End-user and developer support for geWorkbench is provided through the caBIG® Molecular Analysis Tools Knowledge Center, a component of the caBIG® Enterprise Support Network.
Graphical User Interface
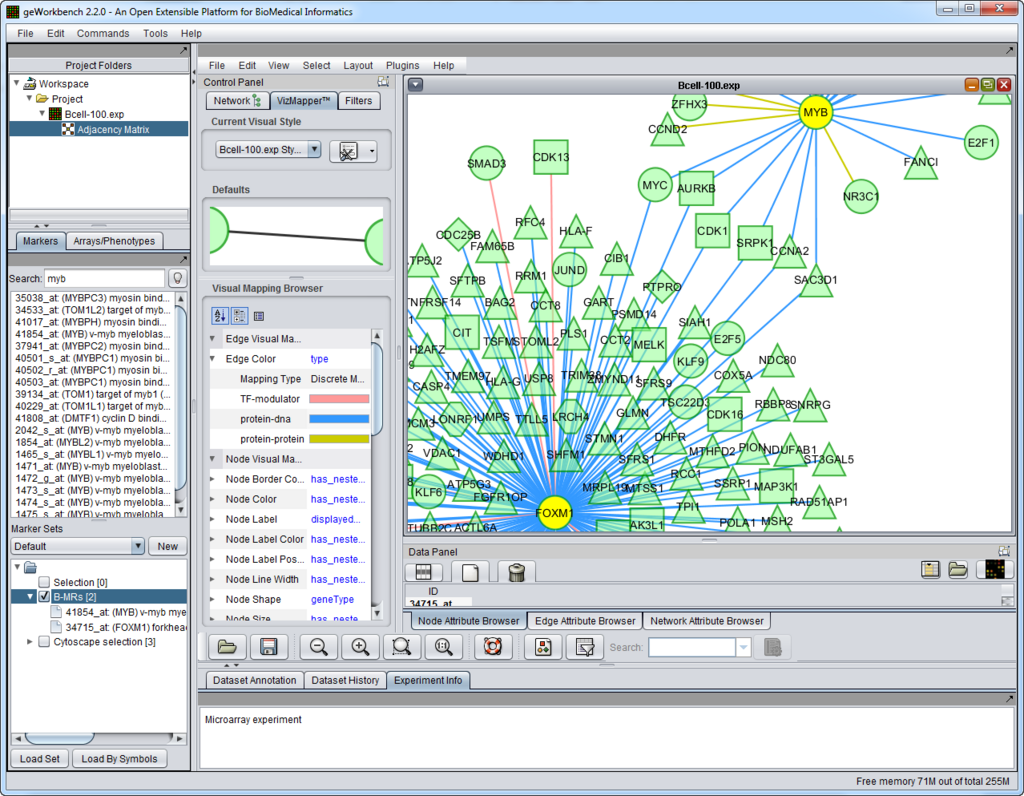
The figure above shows the full geWorkbench graphical user interface. The Cellular Network Knowledge Base B-cell interactome was queried for interactions of two master regulator genes, and the result displayed in Cytoscape.
Summary of changes in geWorkbench release 2.2.0
geWorkbench 2.2.0 is a major release containing more than 180 new features, enhancements and bug fixes. The most important of each are summarize below.
Overview of new features
- New network comparison and manipulation features were added to the Cytoscape component,allowing comparison of CNKB interactomes to other expression datasets, projection of t-test results onto a network, and creation of subnetworks from the result of these comparisons.
- The Gene Ontology component can now serve as a full, standalone GO term browser.
- Options for import and export of interaction networks (interactomes) were added.
- Two new filters were added to give the user options to deal with many-to-many relationships between genes and markers (probesets).
- Two new analysis components were added. One calculates differential expression fold-change, and the second provides a front-end to GenePattern GSEA (Gene Set Enrichment Analysis).
- Improvements were made to the Master Regulator Analysis component - it can now accept any user-supplied list of "signature genes", and the "bar-code" display was altered to match the style used in recent publications.
- A new Social Center feature was added to the genSpace component, allowing users to directly interact with each other. Improvements in workflow
saving and visualization were also made.
- A major code refactoring was completed, simplifying a number of core data structures to improve performance and prepare for the development of a new web version.
New features in detail
- CNKB
- (#2389) Export complete CNKB interactomes (SIF or ADJ formats). The Cellular Network Knowledge Base (CNKB) stores a number of "interactomes" derived from computational analysis of various types of gene expression data. These interactomes, and other interaction datasets, can be queried in geWorkbench to find targets which interact with genes of interest. With this release, complete interaction networks stored in the CNKB can be exported to a file, in either SIF format or as an ARACNe-format adjacency matrix.
- Cytoscape
- (#2424) Compare differential expression results to an interaction network ("interactome"). This feature calculates the Pearson's correlation coefficient for the expression profiles of each pair of nodes connected by an edge in an interaction network. Only those portions of the network which have a correlation coefficient above a user-set threshold will be displayed.
- (#2424) Create a new subnetwork containing only edges exceeding the calculated correlation threshold.
- (#2429) From an existing network, create a subnetwork containing only nodes in a marker set defined in the Markers component.
- File Parsers
- (#2388) Import an ARACNe adjacency matrix from a file. Either gene symbols or probeset names can be used.
- Filters
- Multiple probeset per gene filter (#2444) - For genes with multiple probesets (markers), retain only the probeset with: (a) highest coefficient of variation, (b) highest mean, or (c) highest median expression across all arrays.
- Multiple Entrez GeneID Filter (#2445) - Filter out markers which are annotated to (a) no Entrez gene id, or (b) multiple Entrez gene ids.
- Fold-change Analysis
- (#2431) This is a new component that performs fold-change analysis and places markers that pass the specified threshold into two new sets in the Markers component, one for markers with positive fold-change, and the other for those with negative fold-change.
- Gene Ontology
- The Gene Ontology component is now always available when a microarray dataset has been loaded along with its annotation file. The GO Tree can be browsed or searched for any term. The markers annotated to any term can be returned to a new set in the Markers component.
- (#1875) The most recent Gene Ontology OBO file is now downloaded automatically from the internet when geWorkbench is started, with the option to instead load a specified OBO file from disk.
- GenePattern GSEA
- A front-end to GSEA running on any GenePattern server has been added.
- genSpace
- A new social networking feature, the Social Center, has been implemented, allowing users to directly interact with friends or create networks (chat and share).
- Users can now create their own Workflow Repository, where they can collect and comment on their favorite workflows.
- The GUIs for Workflow Visualization, Real Time Workflow Suggestion and Workflow Statistics have been updated.
- Master Regulator Analyis (#2523 and others)
- Allow any user-supplied list of markers to be used for the phenotype signature.
- Bar-code graph revised to match style of published work.
Other Enhancements/Fixes
- ARACNe
- (#2366) - In ARACNe, bootstrapping is re-enabled but only single threaded.
- (#2482) - ARACNe results can now be pruned to retain only highest MI edge per gene-gene pair, or return all edges.
- BLAST (#2419) - Continued improvements to BLAST interface to match NCBI website functionality and to improve usability.
- CCM - "Sequence Analysis" has been retitled as "BLAST Analysis", "Alignment Viewer" has been retitled as BLAST Alignment Viewer.
- Gene Ontology Viewer (#2391) - When a marker set is returned for a GO term, the set is given the term name.
- GEO Soft (#2402, #2462, #2465) - GEO Soft parsers improved to handle various special cases - multiple platforms, missing values, mixed sample and data matrix files...
- Grid Services (#1773) - Simplified grid service activation (removed one radio button).
- JMOL (#2505) - Updated to JMOL version 12.0.35.
- Markers/Arrays component (#2430) - Dynamic filtering of displayed marker or array list as search term is entered.
- MarkUs
- (#2500) - Add ability to retrieve prior MarkUs jobs by job id
- (#2509) - Add private key option to MarkUs job submission
- MINDy (#2214) - Corrected sign of modulation effect in table displays.
- Pattern Discovery (#2119) - Corrected display problems in Pattern Discovery related to regular expressions and use of substitution matrices.
- Preferences (#2393) - Added ability to reorder data sorted by marker name, gene name, or original order (set in preferences, affects all components).
- Sequence Retriever (#2518) - Fixed problem with obtaining name of latest human genome build from Santa Cruz.
- Tabular Microarray Viewer(#2253) - Tabular Microarray viewer now allows adjustable precision in display, and choice of fixed or scientific notation.
- t-test (#1626) - Changed math package used in order to correct precision problem with p-value calculation at very small p-values.
- Volcano Plot (#2492) - extreme point color range corrected.
Changes to documentation (Tutorial sections)
Note - not all of the new features described above have been documented as of the release date, but they will be added as quickly as possible to the tutorial chapters on this website.
- ARACNe
- Corrected and improved descriptions of DPI Tolerance and DPI Target List.
- Added Technical Notes section and usage notes.
- Mapped relationship of ARACNe command line options -s and -l to ARACNe in geWorkbench.
- Rewrote introduction.
- CNKB - all screenshots on tutorial page updated to reflect
- Interactome and version details display.
- Illustration of new display options in Cytoscape (see below).
- Cytoscape - Added new section to show all new features for release 2.2
- Correlation Overlay,
- t-test overlay,
- subnetwork creation,
- interaction type coloring etc.
- Data Subsets - Arrays
- Many new and revised screenshots added.
- Added new full section on visual properties editor.
- Now call the collections of sets "Lists" (pulldown menu entries).
- Much material was previously in a separate "Examples" section; this has now all been moved into the primary description of each menu item.
- Data Subsets - Markers
- All the same changes made to "Data Subsets - Arrays" were also made to "Data Subsets - Markers".
- Material unique to the Markers component was added as needed.
- File Formats - Error dialog offering 3 choices of action when duplicate entries are encountered in an annotation file (#1624).
- Local Data files - full update, including
- updated descriptions of GEO files.
- updated screenshots and descriptions,
- added additional material about file browser, merging, annotation files....
- Menu Bar - New tutorial written to cover all actions available in the top level menu bar.
- Pattern Discovery - all screenshots replaced. Changes reflect:
- Fixed display of regular expression matches in Full Sequence view after running with "Exact" unchecked.
- Better illustration of multiple pattern displays in viewer.
- In Advanced tab, matrix and threshold settings now disabled when "Exact" is checked.
- Projects
- Improved descriptions and added additional screenshots throughout
- Added missing options such as RCSB PDB.
- Add new section on Workspaces
- SVM - all new chapter written. It is updated to reflect small changes to the Test tab GUI.
- t-test
- Fully revised all sections.
- Added sections on Volcano Plot and Color Mosaic
Previous Releases
See the Previous Releases page for full details.