MARINa
Contents
Overview
MARINa will run the gene set enrichment analysis (GSEA) algorithm for each transcription factor (TF) on the chip to calculate if the regulon of a TF is enriched for genes that are differentially expressed among the 2 classes of microarrays. Following all GSEA runs MARINa will also perform an additional "shadow" analyses.
Inputs
- Network - the network (e.g. from ARACNe) upon which MARINa will operate.
- GSEA and shadow/synergy analysis run parameters:
- Minimum Number of Targets: minimum number of genes in a regulon for a GSEA run (a positive integer).
- Minimum Number of Samples: minimum number of arrays in the input microarray set in order to run sample-based permutations in GSEA (a positive integer).
- Number of GSEA permuations: number of GSEA permutations to compute enrichment score distribution (a positive integer).
- GSEA Tail: whether 1-tailed (1) or 2-tailed GSEA (2) is to be performed. If all correlations in the network file are greater than zero, then one-tail GSEA will be performed regardless of the value the user assigns to "tail".
- Shadow P-value: significance threshold for shadow analysis.
- SynergyP-value: significance threshold for synergy analysis.
- Retrieve prior result with ID (checkbox) - if checked, all parameter fields are disabled and the prior ID field is enabled. The user can retrieve the results of a previous run.
other input
- GSEA P-Value: This parameter appears on the "Main" parameters tab. It indicates the enrichment score p-value below which a regulon is considered enriched in differentially expressed genes.
set by program
- paired: boolean value indicating if the microarray samples should be treated as paired or not.
Network alternative 5-column file format
MARINa in geWorkbench will normally be set up to work with a network that is either already in the Project area, or that is loaded from disk in a standard format, e.g. an ARACNe-style adjacency matrix or SIF file. geWorkbench will generate a file in the following 5-column format which is then sent to the remote MARINa service. However, a file in this format can also be directly loaded into the MARINa component.
Each line represents a network edge and comprises five tab-delimited columns:
- Transcription factor id: A string that provides the transcription factor end of the edge. This is usually a probeset id or a gene symbol.
- Target id: A string that provides the target end of the edge. This is usually a probeset id or a gene symbol
- Mutual information: The mutual information (MI) of the edge (a real number). If the edge MI is not known/available, the value 1 can be entered here.
- Spearman's correlation: The Spearman's correlation for the edge, computed on the original microarray set that gave rise to the ARACNe network (a real number). If not known/available, the value 1 can be entered here.
- P-value for Spearman's correlation. The p-value associated with Spearman's correlation found in the previous column (a real number between 0 and 1). It this p-value is not known/available, a value of 0 can be entered here.
- Microarray set file. Provides the expression data that MARINa uses for calculating differential expression. Data in this file are given in the .exp format used by geWorkbench ( here are the first five lines from a sample file).
- Class 1 (cases) and Class 2 (controls) files. The arrays that comprise the 2 classes which are used for the calculation of differential expression are provided in 2 files, listing the arrays in each of the 2 classes. Here are 2 such example files: resistant_cellines.txt and sensitive_cellines.txt. In each file, every array is listed in a separate line.
Running MARINa
Results
With sample shuffling:
With probe shuffling:
The MARINa results file contains one line for each TF whose GSEA enrichment score renders it significant above the user-specified p-value threshold (pvalue_gsea), as long as such TF is not "shadowed" by other significant TFs. NOTE: for the time being, we will ignore below (and in our implementation of the grid service) the results of the synergy analysis.
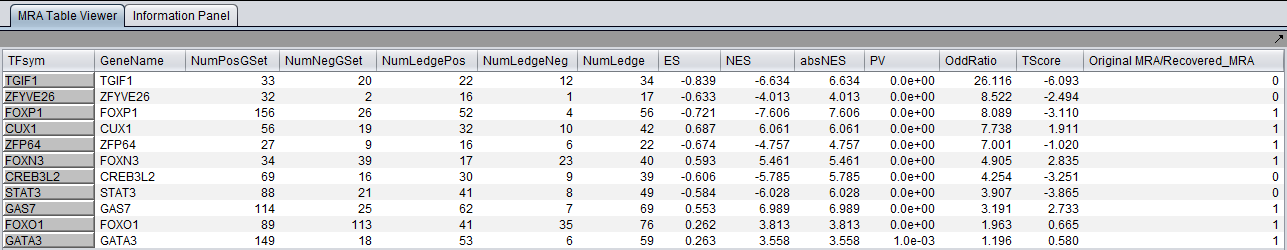
The columns represented in the results are as follows:
- TFsym: transcription factor id (usually this is the probeset id).
- GeneName: transcription factor gene name.
- NumPosGSet: number of genes (among those in the regulon of the transcription factor) which are positively regulated by the TF. Only genes represented in the microarray set are counted.
- NumNegGSet: number of genes (among those in the regulon of the transcription factor) which are negatively regulated by the TF. Only genes represented in the microarray set are counted.
- NumLedgePos: number of genes (among those counted under the NumPosGSet column) which are in the GSEA leading edge.
- NumLedgeNeg: number of genes (among those counted under the NumNegGSet column) which are in the GSEA leading edge.
- NumLedge: sum of NumLedgePos and NumLedgeNeg.
- ES: GSEA enrichment score for the regulon of the TF.
- NES: GSEA normalized enrichment score for the regulon of the TF.
- absNES: absolute value of NES
- PV: p-value of normalized (?) enrichment score.
- OddRatio: (NumLedge/(microarray set genes in the regulon of the TF))/((number of differentially expressed genes left of the leading edge)/(total number of microarray set genes)) - <Aris>please check with Mukesh that this is the correct definition</Aris>.
- TScore - t-test t-statistic.
- MeanClass1: mean expression value of TF among all "Class 1" arrays.
- MeanClass2: mean expression value of TF among all "Class 2" arrays.
- Note - there are no MeanClass1 and MeanClass2 when probe shuffling is performed.
- Original MRA/Recovered_MRA: a value of 1 in this column means that the TF was found to be enriched by GSEA (above the significance level pvalue_gsea) and that it was not shadowed by any other TF. A value of 0 means that the TF was found to be enriched by GSEA and that it was shadowed by another TF and that it remained enriched even after the common targets with the other TF were removed from its regulon.
Extensions for the geWorkbench MRA component
To invoke the MARINa service from within the geWorkbench MRA component (and display the analysis results) we should make the following changes:
MRA Analysis component
- MARINa. In here we specify the params that are specific to the MARINa analysis.These are:
- The Class 1 (i.e. case) and Class 2 (i.e. control) arrays. These should be selected (activated) in the Arrays component available array sets.
- Note the two selected array sets have no arrays in common. An error popup should notify the user if either of these conditions are violated.
- The values for the applicable parameters listed under "GSEA and shadow/synergy analysis run parameters" above.
- "paired", - if the user only includes one class (control), then it is paired. If both classes are provided, it is unpaired.
- The Class 1 (i.e. case) and Class 2 (i.e. control) arrays. These should be selected (activated) in the Arrays component available array sets.