Difference between revisions of "Tutorials"
| Line 5: | Line 5: | ||
geWorkbench is designed from the ground up to be extensible. New modules can be programmed to interact directly with its framework, or existing code can be wrapped in a geWorkbench adaptor to allow seamless communications with the framework and other modules. | geWorkbench is designed from the ground up to be extensible. New modules can be programmed to interact directly with its framework, or existing code can be wrapped in a geWorkbench adaptor to allow seamless communications with the framework and other modules. | ||
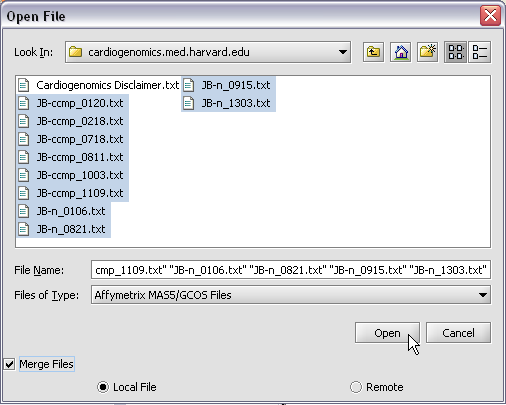
| + | To start using geWorkbench, one must supply initial datafiles. For microarray data, several formats are currently available, including MAS5/GCOS text files, GenePix files, and a simple, geWorkbench-specific matrix format. In the next section, we will show how to read in MAS5 format files and write out a matrix file. For sequence data, fasta format files are accepted. | ||
| + | |||
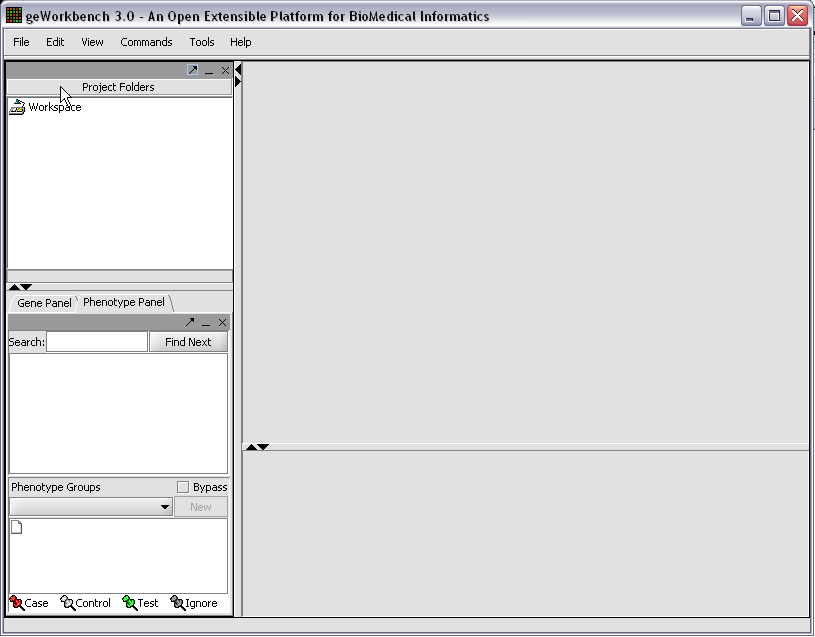
| + | When first started, geWorkbench appears so: | ||
| + | |||
| + | [[Image:T_StartupState.png]] | ||
| Line 11: | Line 16: | ||
| − | + | ||
| − | [[Image: | + | [[Image:T_NewProject.png]] |
| + | |||
| + | |||
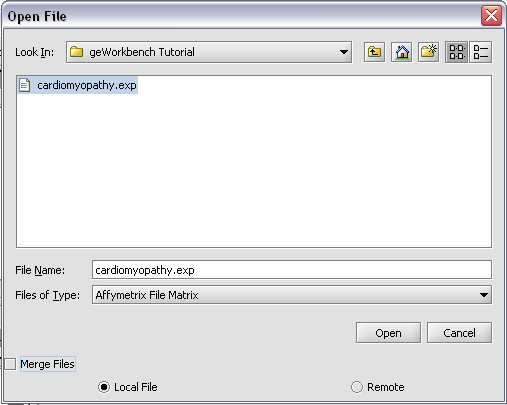
| + | [[Image:T_OpenFiles.png]] | ||
| + | |||
| + | |||
| + | [[Image:T_SelectMAS5.png]] | ||
| + | |||
| + | |||
| + | [[Image:T_Chip_Type_Message.png]] | ||
| + | |||
| + | |||
| + | [[Image:T_MAS5_display.png]] | ||
| + | |||
| + | |||
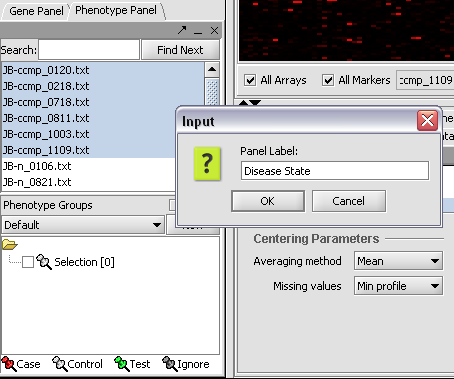
| + | [[Image:T_PanelLabel.png]] | ||
| + | |||
| + | |||
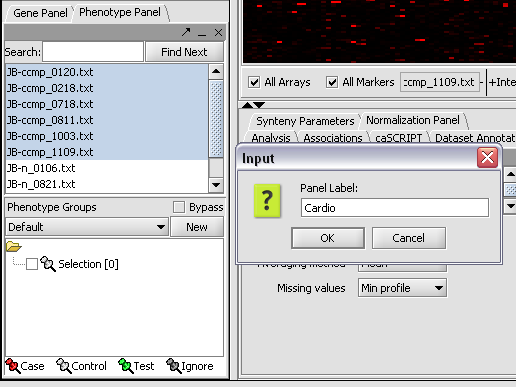
| + | [[Image:T_PanelLabelCardio.png]] | ||
| + | |||
| + | |||
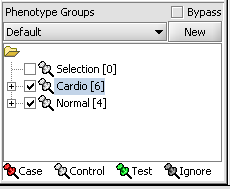
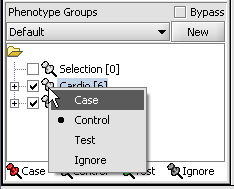
| + | [[Image:T_PhenotypesPriorToCase.png]] | ||
| + | |||
| + | |||
| + | [[Image:T_PhenotypeSettingCase.png]] | ||
| + | |||
| + | |||
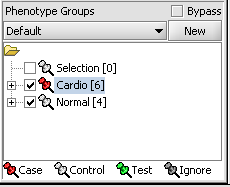
| + | [[Image:T_PhenotypeCaseSet.png]] | ||
| + | |||
| + | |||
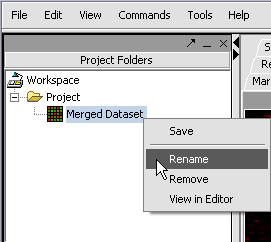
| + | [[Image:T_RenameDataset.png]] | ||
| + | |||
| + | |||
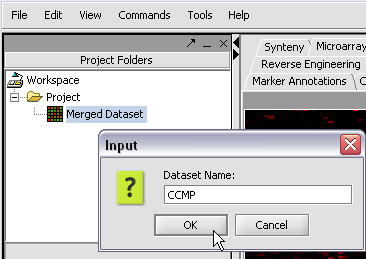
| + | [[Image:T_RenamingDataset.png]] | ||
| + | |||
| + | |||
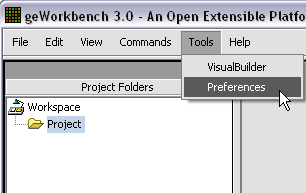
| + | [[Image:T_ChangePrefs.png]] | ||
| + | |||
| + | |||
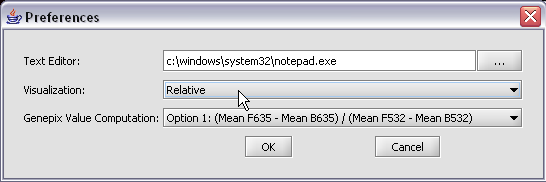
| + | [[Image:T_ChangePrefsToRelative.png]] | ||
| + | |||
| + | |||
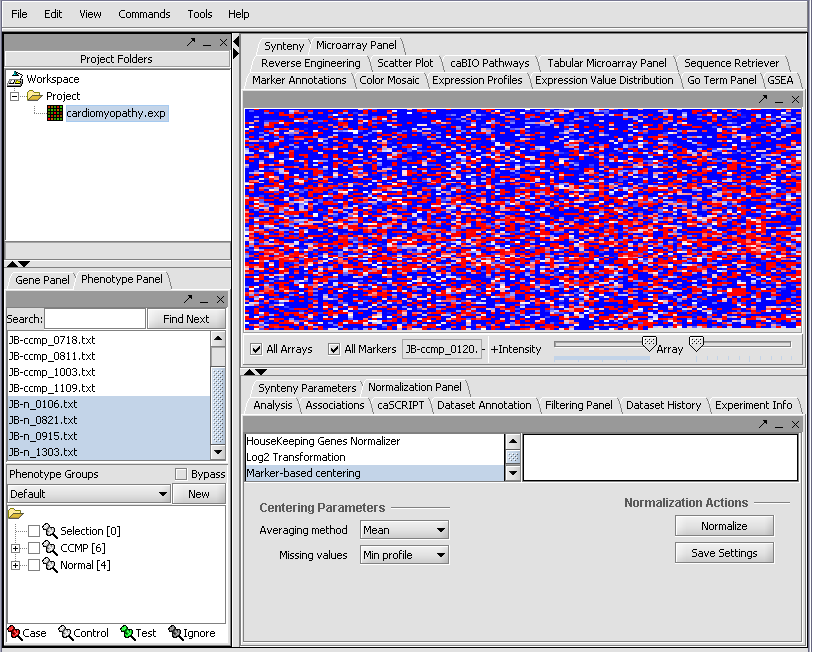
| + | [[Image:T_OpenCardio.png]] | ||
| + | |||
| + | |||
| + | [[Image:T_RelativeDisplay.png]] | ||
Revision as of 19:31, 27 January 2006
I. Getting Started
With geWorkbench you can work with both mircoarray gene expression data and with gene or protein sequences. Many kinds of analysis are supported - for microarrays, there are filtering and normalization, basic statistical analyses, clustering, network reverse engineering, as well as many common visualization tools. For sequence data there are routines such as BLAST, pattern detection, transcription factor mapping, and syntenic region analyis. Furthermore, genomic sequences around markers of interest found in microarray experiements can be easily retrieved and, for example, used for promoter/TF analysis.
geWorkbench is designed from the ground up to be extensible. New modules can be programmed to interact directly with its framework, or existing code can be wrapped in a geWorkbench adaptor to allow seamless communications with the framework and other modules.
To start using geWorkbench, one must supply initial datafiles. For microarray data, several formats are currently available, including MAS5/GCOS text files, GenePix files, and a simple, geWorkbench-specific matrix format. In the next section, we will show how to read in MAS5 format files and write out a matrix file. For sequence data, fasta format files are accepted.
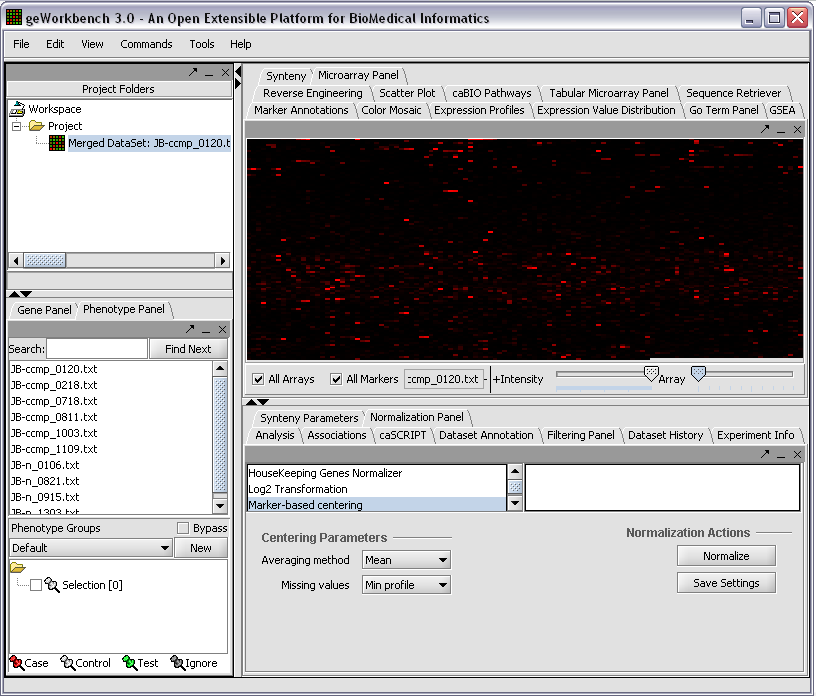
When first started, geWorkbench appears so:
II. Loading Data