Difference between revisions of "User:Floratos/MRA"
(→Prerequisites) |
(→Parameters and Settings) |
||
| Line 19: | Line 19: | ||
==Parameters and Settings== | ==Parameters and Settings== | ||
| − | === | + | |
| − | + | ===Load Network=== | |
| − | === | + | There are 2 ways to designate the interaction network that will be used for computing the neighbors of the candidate master regulator genes: |
| − | === | + | * '''From File''': by choosing a file that describes a network (see example and format description below). |
| + | * '''From Project''': by selecting a node from the project folders which represents an interaction network. Several analytical components in geWorkbench (e.g., [[Tutorial_-_ARACNE | ARACNE]]) produce results nodes that can be utilized for this purpose. | ||
| + | |||
| + | ===Transcription Factors=== | ||
| + | There are 2 ways to designate the candidate master regulator genes to use (in most cases such genes will be transcription factors, hence the explicit use of the term in this field): | ||
| + | * '''From File''': by choosing a file that contains a list of (comma separated) marker names. | ||
| + | * '''From Sets''': by selecting one among the marker sets within the “Markers” component. | ||
| + | |||
| + | ===T-test p-value (alpha)=== | ||
| + | Differential expression between the 2 phenotypes of interest is assessed using a t-test. The p-value provided by the user indicates the significance threshold below which a gene’s average expression is presumed to be significantly different in the 2 sets of arrays. Additional parameter settings affecting the execution of the t-test are defined within the “T-test” subtab. A description of the meaning of these parameters can be found in the [[Tutorial_-_Differential_Expression | “Differential Expression”]] tutorial. | ||
=Working with and viewing the analysis results= | =Working with and viewing the analysis results= | ||
Revision as of 16:39, 19 October 2009
|
Aris's Home | Enhancements | Release v1.1 | Bison Issues | MRA | Release Testing | Home Page |
Contents
Overview
Regulatory activity in the context of specific cellular phenotypes can be modeled using interaction networks. These are graphs where nodes represent genes and an edge between two nodes A and B means that genes A and B are participants in the same regulatory activity. E.g., A can be a transcription factor for B; or, A can be an miRNA that silences B. Analysis of such regulatory networks [refs] has convincingly demonstrated their scale free nature which is dominated by a relatively small number of nodes with a large degree of connectivity. The genes corresponding to those nodes are known as "master regulators" and collectively orchestrate the regulatory program of the underlying cellular phenotype(s).
The master regulator analysis (MRA) component in geWorkbench combines regulatory information from interaction networks with differential expression analysis. The objective is to place differentially expressed genes within a regulatory context and identify the master regulators responsible for coordinating their regulation, thus highlighting the regulatory apparatus driving phenotypic differentiation. Specifically, given an interaction network I, a master regulator gene A, and two sets of microarrays representing two distinct phenotypes, MRA computes the intersection between two sets of genes:
- The neighbors of A in the interaction network I.
- The set of differentially expressed genes in the array data from the two phenotypes of interest.
Fisher’s exact test is then used to quantify how likely it is to encounter an intersection of the observed size by chance alone. A small p-value is taken to imply that gene A may play a significant role in controlling the regulatory program that leads to the differential phenotypes.
Setting up an MRA run
Prerequisites
- First confirm that the MRA component is available in geWorkbench. If not, it can be loaded using the Component Configuration Manager.
- The MRA will be listed along with the other analysis routines within the geWorkbench Analysis pane.
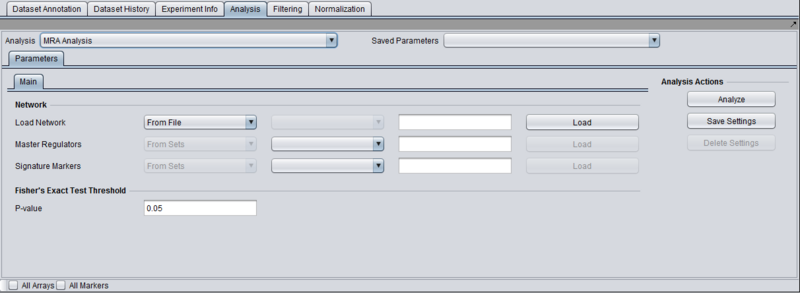
Parameters and Settings
Load Network
There are 2 ways to designate the interaction network that will be used for computing the neighbors of the candidate master regulator genes:
- From File: by choosing a file that describes a network (see example and format description below).
- From Project: by selecting a node from the project folders which represents an interaction network. Several analytical components in geWorkbench (e.g., ARACNE) produce results nodes that can be utilized for this purpose.
Transcription Factors
There are 2 ways to designate the candidate master regulator genes to use (in most cases such genes will be transcription factors, hence the explicit use of the term in this field):
- From File: by choosing a file that contains a list of (comma separated) marker names.
- From Sets: by selecting one among the marker sets within the “Markers” component.
T-test p-value (alpha)
Differential expression between the 2 phenotypes of interest is assessed using a t-test. The p-value provided by the user indicates the significance threshold below which a gene’s average expression is presumed to be significantly different in the 2 sets of arrays. Additional parameter settings affecting the execution of the t-test are defined within the “T-test” subtab. A description of the meaning of these parameters can be found in the “Differential Expression” tutorial.