User:Rfriedman
Functionality Comments
Rich, add functionality comments and new feature suggestions here.
One quick inital suggestion. geWorkbench should be able to import files in the following GCG formats: sequence, mutiple sequence, and rsf.
(3/23/06) A more robust couterpart of k-means clustering with statistical estimates for micorarray analysis is described in the following papers:
3/30/06 I don't like the slider to change arrays in the microarray widow. The identity of an array is a fixed, not a variable quantity. I suggest that a pull-down window for this would be better.
4/7/06 I suggest asking "are you sure" when a user asks to remove a project. 4/26/06 It would be very helpful if the workbench could display an hourglass, or a watch,
or a sundial or something, when it ia loading or working - for example when it
is loading micorarray files from a remote database.
5/25/06. I just installed Version 1.03. In the Windows menu it says version geWorkbench 1.0 and on top of the geWorkbench GUI its says geWorkbench 1.0. I suggest that
all labels give the full workbench version.
5/25/06 The two tutorial sets should be included in the download automatially.
5/25/06 I would like to ammend my recommendation of 4/26/06 to inlcude an estimate of the time a task will take, so that people may use it more easily.
5/25/06 When I spoke to the group, Ken had stated that the intensities in the microarray viewer did not correspond to an image of the chip. In which case the phrase "microarray viewer" is misleading. In fact I am not sure to what the intensities an spacing in microarray viewer corresponds.
5/25/06 I think that "Get bioassays" is a poor command on 2 grounds: 1. I am not used to "bioassays" being used in place of "arrays" or "array data". 2. We are obatining a list, rather than loading the bioassays into the program. What I think we eman then is "list arrays".
Additionally, it is not clear what format the arrays are being loaded (Cel, normalized probeset intensities, etc).
5/25/06 Some indication that a work is in progress should be given while the arrays are being loaded.
5/25/06 I suggest that a dummy new source be made available to the users to learn how to access a remote source and I suggest that intructions for posting a remote source be made available. Doing these things will increase the ease with which users can use the workbench in collaborative projects.
5/30/06 The terms "marker" and "phenotypes" are not optimal. In the microarray world we use "probsets" (affymetrix) or "probes" (glass-slides) instead of "markers". "Arrays" is much more informative than "phenotypes" because there can be several arrays for a phenotype, or arrays can represent different patients rather than a phenotype, or because arrays can correspont to points in a time series. Also, you might want to reserve "phenotype" for instantiations that have precise defintions in a controlled vocabulary.
5/31/06 With respectto the tabular microarray view. There is also a "probe number" for affy chips (1,2,3. ..) based upon its poistion in a sort. It would be useful to have a colum for that. It would also be useful to have seperate, searchable columns for the following 3 items: 1. Probe id. 2. Gene name. 3. Gene defintion.
(If it sounds as if I am thinking of Excel here - I am).
5/31/06. I strongly recommend that there be a way to reverse filtering, by a global undo command or some other means, so that the user may try different filters.
6/14/06 Inclusion in the announcments mailing list should be made an integral part of teh downloading process.
6/14/06 The "expression thresshold filter" instructions shoudl be clearer. stating "Filer values insdie range" is ambiguous in that it is not clear if those values are left after filtering or removed by filtering (I believe that the later is the case). I suggest the language be changed to "remove values inside range". or "flter-out values inside range".
6/29/06 I recently did some Hierarchical Clustering using Cluster 3.0. Instead of simply filtering by absolulte M value, its also enables the user to retain genes that are larger than a given mvalue in a user-specifiiable
- of experiments. It also offers the following options:
1. %. present >= of chips (this only works if you use present/absent threshholds rather than statistical noise. 2 .SD gene vector >=X to remove genes with insufficient variability. 3. Max-Min >= another variability filter.
I can see why someone might want to use 2 or 3.
7/17/06 Tabular micorarray format - The column widths on the tabular microarray format should be suffcient to accomodate the whole title of the chip.
7/17/06 The color mosiac only makes sense if the data already has a log2 or other variance-stabilization transformation. As is, an unsuspecting user can look at real values at this can be confusing. Furthermore, heatmaps make teh most sense for log ratio comarisons versus a standard.
8/8/06 The tabular micorarray viewer should be saveable as an Excel spreadsheet. all tables should be saveable as an excel spreadsheets.
8/10/06 A linear or spline fit to the reference line in the scatterplot would be helpful.
8/11/06 A good filter feature would be to give the user an option of accepting or rejecting the filtering based upon the number that survived the filtering prior to acceptance. Also, there should be the option of blowing up the heatmap. In general the functionality of cluster 3.0 (written by our own Michiel de Hoon) and JavaTreeview should be reproduce for clustering.
8/14/06 A word about t-tests. It is very common in the microarray field for experimentalists to not give sufficient numbers of replicates to get good statistics. The word in the statistical community is to use some variant of a Bayesian t-test which pools variances of similar sample sizes to take into compensate for small smaple size. This started with Cyber-T, but the 3 most used and validated ones are: 1. LIMMA (LInear Models for Microarray Analsysis) from Gordon Smyth based on earlier work by Terry Speed and Ingrid 2. SAM (Significance Analsysis for Micorarrays) by Tibrishani and coworkers. 3. The method included in BRBArray tools, by Simon, Radamacher, and cowrkers.
I am told that LIMMA and SAM give similar results to one another and that BRBArrayTools gives somewhat different results to the two former programs, but not necessarily inferior results.
GeneSpring and GeneTraffic also have their own versions of this method. My anecdotal experiene is that GeneTraffic does not match the results from LIMMA.
I recommend that some version(s) of the Bayesian method be included in geWorkbench. Both LIMMA and SAM are available as part of Bioconductor and therefore can be ported to geWorkbench as part of a of a general Bioconductor port.
8/14/06 i strongly recommed that the benjamini-Hochberg False discovery rate correction be offered as an option. In fact all of the options in the current version of AffyLmGUI would find use.
8/14/06 I suggest that the heat map ratehr than the volcnaoc plot be the default display onb the t-test output.
8/15/06 With respect to the multiple t-test, an obvious improvement, alluded to in the functionality write-up is to take into account multiple comparisons. The most basic way to do this is to add terms corresponding to the variance of all of the smaple studied in the denominator of the expression for t. An expresion for this appears in the powerpoint presentation that I gave to the geWorkbench development group. This correct is different and more fundamental than additional multiple comparison corrections Bonferroni, Benjamini-Hochberg etc. Indeed doing Benjamin-Hochberg corrections for multiple tests (probesets, i.e. "markers) and multiple phenotypes simultaneously has not been implemented by the microarray statistical community to my knowledge. So I wouldn't worry about it for geWorkbench. However LIMMA, SAM, and BRBarrayTools each has its own version of correction at the level of the t-test, and I suggest that at least the first two be implemented in geWorkbench.
I realize that the above paragraph might be rather cryptic. I am available to discuss the considerations involved with the developemnt team.
9/12/06 It is important that geWorkbench be able to import micoarray data in the 2 formats used by Entrez (GEO) (The Entrez Gene expression Omnibus databse). please see the Entrez site for format information.
9/12/06 I have had aneecdotal experience that if the latest version of Java is installed after geWorkbench is installed there is a problem and geworkbench has to be uninstalled and then reinsatlled to work. Xiaoqing says that this shouldn;t be the case, but I just thought I would let you know what had apparently happened.
9/13/06 I believe that the Workbench's utility woulf be greatly enhanced if it containded menus for submission of sequences to GenBank and Microarray data to GEO. This is especially important in the latter case where the burden of MIAME compliance is considerable. Furthermore, the time for users to specify their MIAME info is when they first read the data into the Workbench, so that the task becomes spread over and not a burden at the very end when the user has to upload into GEO in order for the manuscript to be cleared for publication. The Workbench should remind the user frequently of unspecified and unnotated files.
9/28/06 A much larger assortment of chip-types should be offered.
9?28/06- A general functionality need is the ability to analyze SNP-chips for loss of heterozygosity, copy number, and whole genome associations. The best package for the first 2 is dChip. I am just learning about whle genome association analysis so that I cannot make a recommendation at this time, but expect to be able to soon. We can talk about SNP-chips if and when you are ready to pursue them.
10/5/06 When I opeb webmatrix.exp, it immediately goes to the ARACNE module. This is confusing, because the tutorial has it go to the microarray viewer.
10/11/06 I suggest that the caption accompanying: Microarray Viewer: Filtering: Missing Value Filter, be changed from "Maximum number of Missing Arrays" tp "Remove markers that are missing in [NUMBER BOX] arrays". It would be much clearer.
10/12/06 That the deviation bound is in raw score units should be mentioned
in the GUI. Perhaps, someting along the lines of "Remove markers which
vary by more than [NUMBER BOX] raw scores". A filter based on standard deviation
would also be useful.
10/16/06 I tried the deviation filter again today and am convinced that the standard deviation filter is a more rational way to deviation filtering than the absolute value of the deviation. This is because it is hard to specify a meaningful standard deviation cutoff for markers, because markers vary in absolute magnitude of mean and range so widely. The standard deviation measure on the other hand scales with magnitude of mean and upper and lower boundaries. It is a much more natural choice than absolute range.
4/20/07 I suggest that inthe open files menu there be an "all files option".
4/21/07 1. Put a vertical line on the left of all menu windows with arrows at the top and bottom :
/\
|
|
|
|
\/
If the user doesn't see the arrows he will know to enlarge the window. Or as Ken pointed out, when I sent him the above comment, a scroll bar.
4/25/07 At the risk of sounding picky or avisual the curved around icon doesn't immediately suggest submit to me. How about a green light with the word "Run" under it?
4/25/07 The Blast ouptut does not contain the colored bars denoting similarity that appears in the Blast web-site. Ken writes "I think those are extra services provided by the NCBI Blast website itself, they are not part of the data returned using a remote query." If this problem can be overcome it would make a big difference to users who, as a group, just LOVE those colored bars.
4/27/07 The Option "Columbia BLAST sever" didn't work. I got a "Connection Refursed" error message: I queried the user group. Ken replied that "The Columbia BLAST service is not currently operational. There is no fixed time for its reinstatement. The reason is that the Paracel BLAST machine that we used to provide that search capability failed and was retired. We do hope to set up an interface to our cluster to run those BLAST jobs at some point." I suggest that the option of the Columbia service be removed and not restored unless and until there is actually a service corresponding to it". In the meantime users can use the NCBI service either through the web, the geWorkbench interface, or the GCG Netblast program on Cancercenter, which is especially suitable for running many successive Blast jobs as part of scripts. I set up custom databases for Blast, fasta, and Smith-Waterman searches on cancercenter at user request.
5/01/07 I have it set to BLAT. The text book that appears when the mouse is over the start icon is "Start Blast search" (not blast seach). Icon
5/01/07 All of the non-blast functions (BLAT, HMM, Other Algorithms) under sequence comparison should be removed because the functionality is not available). The Columbia server option should be removed from the BLAST menu for the same reason.
5/04/07. The start (curly arrow) and stop(rectangular sign) are in different places in different menus. In pattern recognition tehy are on the top of the page. In BLAST they are at the bottom of the page. A more uniform look would be helpful.
7/12/07 1.06 SPLASH does not have a user-entered Z-value cutoff so that the user is at the mercy of the system as to how many values are displayed.
?12/07 1.06 the help pages should be searchable.
7/12/07 1.06 I suggest that there be better ways to save patterns. Either, just patterns or patterns highlighted in sequences.
7/13/07 1.06 I think that the Globus check box should be removed until such time that Globus is available.
7/18/07 1.06 Having "exact only" as a default checkbox in the advanced pattern discovery is not a clear way to indicate the difference between exacr patterns and the use of a matrix. I suggest that this be part of a scroll down menu in basic and "exact" match be an option along with the similarity matrices.
7/26/07 1/06I am having have had trouble reproducing the finding in the first SPLASH paper that the pattern for 209 H1 histones is
G.S...[ILMV]...[ILMV]
in using the database of 208 histones that comes in the data section I cannot get a single pattern that hits all of them. However with support =100% min tokens=4 density window=12 density tokens=4 Blosum50 similarity threshhold 2 Exact onluy count sequences
I get 3 patterns each of which contains but is larger than the one in the paper: [NDE][RK].G.S...[ILMV]...[ILMV] 1.21E+87 [NDE]..G.S...[ILMV]...[ILMV] 8.21E+42 [RK].G.S...[ILMV] 3.90E+10
7/26/07 1.06 I just realized that the Z-score cutoff was supposed to be setable, and was at some point in the past, because it appears as such in the tutorial.
7/26/07 1.06 The N(26gaps)DRY pattern did not appear on my screen from the GCPRs, perhaps
because its expected Z value from the paper was -11.13 and the top patterns had Zs ranging
from 1.96E138 to 2.34E153.
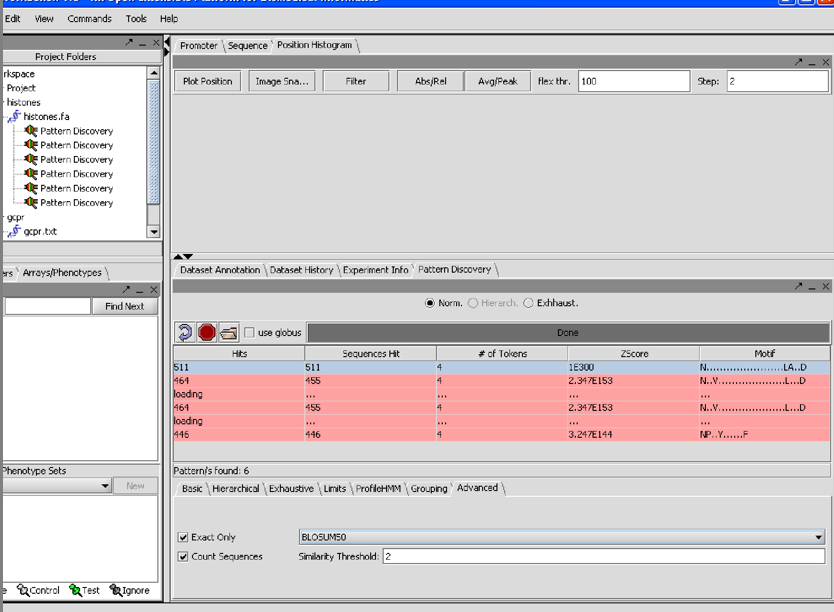
Still, conscientious perspective users will try to duplicate the results in the paper.
7/26/07 1.06 The term "count sequences" does not make a difference in how Splash runs or not and its meaning is unclear looking at the interface. I suggest that it be replaced my an option that tells one how to measure the total number of occurences rather than the percentage. indeed, the user should also be able to specify support in terms of number of sequences, not just % of sequences.
7/26/07 1.06 Splash output often comes out blank. That is to say the boxes say "loading" until I click them. is there anything that I can do on my end about this?
7/26/07 1.06 In the advanced box for pattern discovery there is a scroll bar with the options:
BLOSUM50 BLOSUM100 BLOSUM150
BLOSUM 50 is a similarity matrix based upon the frequency and co-occurrence in alignments of residues in short gapless blocks obtained from aligned proteins of 50% or less sequence identity.
BLOSUM 100 is a similarity matrix based upon the frequency and co-occurrence in alignments of residues in short gapless blocks obtained from aligned proteins of 100% or less sequence identity (All proteins).
So, presumably, BLOSUM 150 is a similarity matrix based upon the frequency and co-occurrence in alignments of residues in short gapless blocks obtained from aligned proteins of 150% or less sequence identity (All proteins). If so, since the maximum possible sequence identity is 100%. how does BLOSUM150 differ from BLOSUM100?
I think there is a mistake here.
Ken subsequently verified that only BLOSUM50 works, and that the development team will either remove the others OR add capability of using real ones.
7/27/07 1.06 I suggest that SPLASH be available both as a command-line open source code ans as a web-server as well as through geWorkbench. I realize that this suggestion might seem to run contrary to the Integrative Genomics Platform philosophy but I believe that just getting a pattern out of a group of sequences should not require learning to use and install the workbench. The strength of the workbench lies not just in its separate applications, some of which, like Splash are not available elsewhere, but in its ability to combine and reuse data of different types. For example the use fo SPLASH to search a blast derived dataset. It is this kind feature that can be stressed in the workbench. I propose that making Splash available through the web and as a standalone Unix command line application would increase the demand for the workbench because people will then want to use Splash in conjunction with other tools. For MAGNet grant renewal purposes, it would be helpful to be able to list the number of citations of MAGNet tools in the literature. The availability of Splash via a command line and a web interface will increase this number of citations.
7/27/07 1.06 I suggest that Postit style expalnations of parameter functions appear as the mouse scrolls over the interface.
8/2/07 1.06 Exhaustive saearch. I suggest that the non-functioning input features be removed an only be restored when they are funcional.
10/19/07 1.06 Promoter. This is a good tool but it might not be state of the art. The emphasis in promoter searching has shifted to using methods of finding conserved and reducing the noise by limiting the search to conserved regions. Here are some web-sites that do this: http://asp.ii.uib.no:8090/cgi-bin/CONSITE/consite/ http://burgundy.cmmt.ubc.ca/oPOSSUM/ http://bioinformatics.wustl.edu/PAP (currently not available)
Another point is that these tools search a databases of sites, not a few selected sites.
Another point is that it would be bettwer if the program to take gene names as input and found the promoters as other programs do.
12/21/07 The graph that shows the expression and relative expression of several markers is extremely useful. However,
it would be more useful if array labels, rather than just
numbers. were given on the Y-axis.
12/21/07 1.06 For color mosiac the tutorial states: "The buttons Pat, Abs, and Ratio, are not currently used". I suggest that they be removed from the display until, which time, if any, they are removed, so as to minimize confusion".
12/24/07 1.06 When I apply the missing value filter the hourglass goes on and off. It would be nice if it were continuous.
12/26/07 1.06 Differential expression. I suggest that the method by which the Bonferroni correction is adjusted be shown in the menu.
12/26/07 1.06. the user currently has a choice of either equal variance or unequal variance t-tests. This can be distinguished by the test for equality of variances (Bartlett test).
12/16/07 1.06log2 transform I got an error message: This data contains non-positve data points. This should not be with raw mas5 data.
Tutorials Comments
Tutorials comments go here.
The initial download should come with all of the datasets in the tutorial (the cardio set was missing when I installed) OR the tutorial should show where these can be downloaded.
3/30/06: Some mention of what the micorarray viewer does should be included in the manual - i.e that it shows a raw image of the chip.
3/30/06: What it means to merge microarray files should be stated more explicitly.
4/07/06 That the chip recognition message is only shown once should be stated. Alternatively maybe it should be shown each time - but not require an okay button.
4/10/06 How to save a merged affy dataset so that one may open it again shoudl be described more clearly. The following points (courtesy of Ken) should be mentioned (and illustated).
1. The set should be saved with an exp suffix.
2. The set can be reopened with the filter set to "Affymatrix Matrix file".
4/24/06 The tutorials comments for opening a remote site are misleading. It should state: 1. Go is clicked for getting the list of micorarray experiments. 2. "Get Bioassays" is necessary for getting a list of arrays in the
experiment-not for retreival.
3. "open" will retrieve the selcted bioassays. I found this veyr hard to use and required correspondence with Kem and a visit from Xiaoqing in orfer to learn to use it.
4/26/06 I suggest that the tutorial not mention adding a new site for remote downlaod umtil such sites are commonly available. Otehrwise it just begs questions from the reader/
5/25/06 I suggest that the tutorial pages state to which version of geWorkbench they apply. This is implicit in the label of the window, that appears in the screenshot, but it should also be on the web-page that the user unloads.
5/25/06 I suggest that the tutorial pages be downlaodable as a pdf file.
5/25/06 I suggest that there be a public mailing list where users can be notified of updates.
5/25/06 I suggest to what the intensities and layout on the microarray viewer slide be discussed.
5/30/06 Designating a group of arrays a "case" causes the thumbtack to be labeled red. However, designating a group of arrays as the "control" does not change the color of the thumbtack. I suggest that the color of the thumbtack be changed to green to distinguish it from a group whose nature has not been demonstrated. Also, the designation "case" is used in clinical and epidemiological research. The corresponding term in laboratory research is "experiemnt".
6/13/06 It should be explained that the microarray viewer image is in probset order split across each row and is not an actual image of the slide.
6/14/06 Examples of each the different filter options should be given in the tutorial.
7/17/06 I believe that you are doing the person learning to use geWorkbench a disservice by showing the heat map instructions in the tutorial before you have shown log transformation (or at least am assuming that there is no log trandformation because some of the numbers are so high). Heat maps are most useful relative to a standard and hence this should be used as part of a didactic example in which a log2 ratio standard is used.
7/17/06 I suggest that the instructions in the tutorial for using the scatter plot graph be more detailed and step-by-step.
8/8/06 I suggest that the difference between a project and a workspace be spelled out.
8/10/06 I suggest the tutorial note explicitly the meaning of the buttons necessary to display the heat map in the micorarray display panel.
8/10/06 There should be some discussion as to where array/phenotype labels come from in the tutorial.
8/10/06 I suggest that there be a tutorial example to plotting the array with a subset of a few probesets.
8/10/06 An illustration of using reference line in the scatterplot would be helpful.
8/14/06 A discussion of the interpretaion of heat maps and volcano plots which wouldbe helpful. I would especially appreciate this in the case of volcano plots, because although I can read the axes, I don't really know how to interpret them. Also an example in which filtering by p-value and by fold change should also be given.
9/14/06 It would helpful if the installation instructions would specify that updates of Windows sometimes includes an earlier version of Java than 1.5 and hence should be checked. It should also state that installing geWorkbench followed by Java 1.5 does not work. It should be specifically stated that the Jave insatllation proceed the geWorkbench installation and that if by mistake, Java is installed after geWorkbench, geWorkbench should be uninstalled and then reinstalled in order to work.This cautionary is consistent with a recent ptoblem I have had and its solution.
9/28/06 A picture of selecting the chip-type should be included in the tutorial which deals with the uploading wo wed-matrix2.
10/5/06 A before-and-after picture of the microaarry viewer application of each filter type to the data and a discussion of the color codes and the reasons for applying the filtering would be helpful. This discussion should include screenshots of the table showing the affect of the filtering and the change in the number of genes.
10/11/06 TI suggest changing" Discards all markers that have missing measurements in at least N microarrays, where N is set by the user" to "keeps all markers that have missing measurements in N or fewer arrays where N is set by the user", in order to bring the tutorial into agreement with what the program actually does and what is stated in the actual GUI.
10/12/06 I suggest that an explict demo of the deviation filter be given in the tutorial.
10/16/06 I suggest that the explict demo of the deviation filter include before and after shots of the tabular microarray viewer and include both absolute and standard deviation options.
4/17/07 I suggest that the tutorails be available on the gforge site.
4/18/07 That retrieving the sequences for a list of markers requires expression data to bebe loaded into the system should be stated explicitly in the tutorial.
4/21/07 Frequent mention should be made in the tutorial that on 15" diagonal screens the user should make sure the menu (otions) window is tall enough to display all of the options and if not, raise the user should raise the Window until they are visible. Please see a complmentary note that I will write today in the tutorials window.
5/3/07 The distinction between Min Tokens and Density Tokens could be clearer. I read the paper so I think I get it, but there should be an example at this point.
5/4/07 Frequent references to the existence of the online help as supplementing the tutorial would be helful.
5/4/07 Create session menu for SPLASH: Tutorial should explicitly state that the port must be set to 80 and that any username will work and that no password is required. On my end a different port number has appeared and it dosn;t work with that.
6/8/07 Version 1.06. When I click "Taxonomy Tree" on the blast output, I don't get anything.
7/12/07 1.06 There should be clear examples as to the meaning of the 3 splash input parameters, with pointers to the variables in the paper. Also, there shoudl be more splash examples. The one given is a good first example, from a technical viewpoint but its biological interest is not meaningful to the user. An example such as the ones in the original splash paper and its sequels will exhibit the power fo the method better.
7/13/07 1.06 This comment applies more to the help pages than to the tutorial per se. The help pages link to the SPLASH pages at IBM. The SPLASH pages at IBM look funny (one long column) when lined to from the help window. furthermore, the pdf files thereby accessed don't come across. in order to read the IBM pages properly, I had to paste teh URL directly into my browser. Since some things have to be explaiend more throughly that is presently the case in the local documentation (e.g. exhaustive discovery) this linking problem presnets an obstacle to use. I suggest that the relevant portiosn of the IBM page be imported directly to the HELP pages.
7/13/07 Since Globus doesn;t work, I suggest that the discussion of it be removed from the help pages, until such time when it does work.
7/18/07 1.06 It would be helpful if the histone example from the first splash paper were to be used as an example in addiiton to the one given.
7/26/07 1.06 The help page for advanced includes the Z-acore which does not appear in the current dialog box.
7/26/07 1.06 the User Guide provides protein-based examples with a clearly labeled test set that comes with the distribution. The web-based tutorial only contains a nucleic-acid based example, which is fine as far as it goes, but which does not fully illustrate the biological power of the program. I suggest that all or part of the User Guide examples be included in the online tutorial. Itr would still be a helpfu, however, l to have the exact same test sets as in the original paper.
7/27/07 1.06 A mooment on the user guide (not tutorial). p. 50 "we will load a database and attempt to discover a common motif in at least 95% of the sequences". However, the example on p. 52 states "Support 80%".
7/27/07 1.06 In my experience a "User Guide" is a general introduction to the conventions of a software application, whereas a "Manual" is an detailed description of the operation of its packages. I think the "User Guide" is really a "Manual" albeit an incomplete one.
7/27/07 1.06 On page 52 of the tutorial, the screenshot shows the sequences displayed with the location represented as a motif. In version 1.06 this only occurs when the higher level Tab that says "sequence" is selected. This is not shown in the user guide.
7/27/07 1.06 I think that the user guide and the online tutorial should be merged. I think that the added detail in the manual really helps orient the user.
7/27/07 1.06 If I am not mistaken the default is exact match. The example in the User Guide will therefore not work as written, because you have to uncheck Exact Only to get it to work.
8/2/07 1.06 The User-Guide on "Exhaustive pattern search" is not very helpful in that it does not state what the parameters mean. Exhaustive searches are not covered in the tutorial document. The section on "exhaustive discovery" under "pattern discovery" in the online databases does not really apply to the current implmetaion in that it treats searching databases selected from a menu rather than a set of sequences loaded into the program.
8/2/07 1.06 In general the user guide should contain everything in the tutorial and the online help. Having to go to 3 differnet docuemnts (plus the IBM splash teatment plus the papers) to figure out what is goin on is a barrier to learning to use the software.
does not really apply
10/19/07 1.06 promoter. Some mention of how the JASPAR Core PSSMs are modified and how the search is done would be informative.
10/22/07 1.06 GO term enrichment. The instructions for loading the marker list in the tutorial are not clear. I loaded the list by clicking "new" under the marker sets and reache the csv file that way. This should be made clearer, step-by-step in the tutorial.
12/14/07 1.06 In the tutorial - dated 8/16/06 Under Arrays/Phenotype se there are now many more sets than are covered in the manual. I suggest that this be updated when the tutorial is revised.
12/14/07 1.06 In the tutorial - dated 3/12/07 Under Viewing microarray datasets, it shows the viewing of a *.cel file. I suggest that a sample *.cel be included in the tutorial dataset and that reading it in be demonstrated in updates of the tutorial.
12/17/07 1.06 I am using color mosaic on the Affymetrix B-cell dataset with GC B-cell and non GC B-cell selected. When I turn the intensity slide-wire some cells appear as green. I was under the impression that this data is the raw data extracted from the cel file with Mas5 without additional centering or normalization. Is this correct? If so, shouldn't all of the values be positive (red)?
12/21/07 1.06 I suggest that a screenshot of the menu that shows the saved image be shown at this point in the tutorial and explicit instructions be given. Otherwise it took me a while to realize that the image was saved.
12/26/07 1.06 Differential expression. I suggested that the tutorial screenshot be updated to replace "class" with "ultrashort designation".
12/26/07 1.06 The tutorila uses the Bonferroni correction, as an exmaple and correctly says that it is the most stringent. It is so stringent that it is generally not used in practice. In practice, the Benjamini-Hochberg FDR is most commonly used. As I have remarked in the comment on the tutorials section, it is not clear if the Benjamin-Hochberg FDR is the one meant by adjusted FDR.
