Difference between revisions of "User talk:Smith"
| Line 2: | Line 2: | ||
| + | When working with microarrays, geWorkBench uses the term '''''marker''''' to refer to a gene probe (in other cases, it can be individual items from other data sets, such as sequences). | ||
Revision as of 22:35, 27 February 2006
stuff cut out from other pages for possible later reuse:
When working with microarrays, geWorkBench uses the term marker to refer to a gene probe (in other cases, it can be individual items from other data sets, such as sequences).
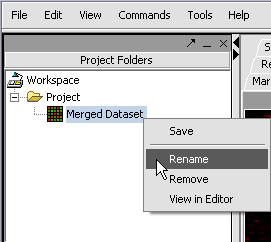
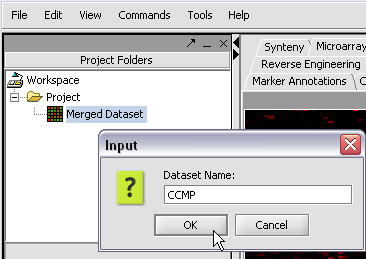
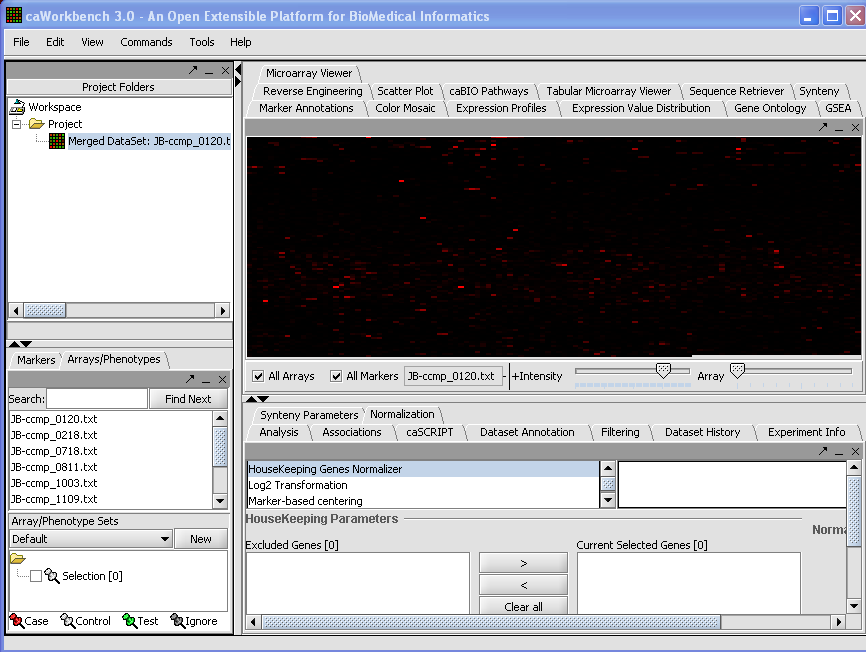
We can also rename the merged dataset by clicking on its entry in the Project Panel.
Here we will call it CCMP.
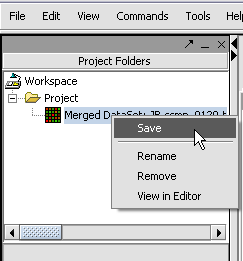
With the datasets merged, classified and named, we can save the dataset for future use. We will call it "cardiomyopathy.exp" (.exp is the default extension for the geWorkbench matrix file type).
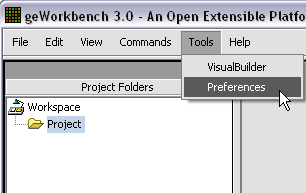
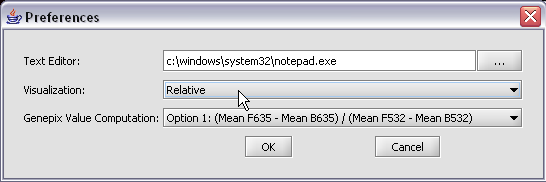
The default display of microarray data is an absolute display. We can change it to a relative display by selecting Tools:Preferences from the top menubar. We have removed the dataset so that we can read it back in using the new preferences.
Here we select the relative display type.
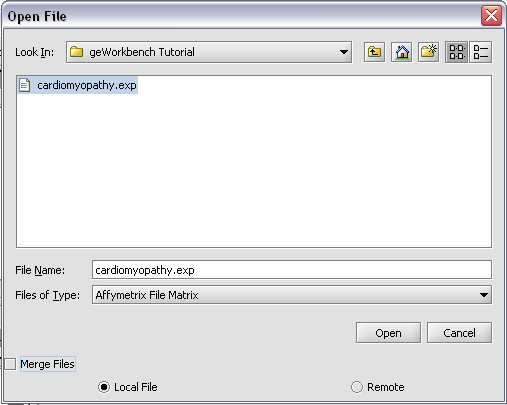
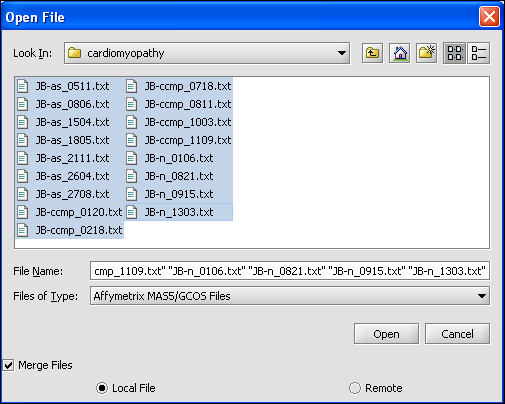
Returning to the Open File dialog as we before by right-clicking on the project entry, we will select the "cardiomyopathy.exp" file we previously saved...
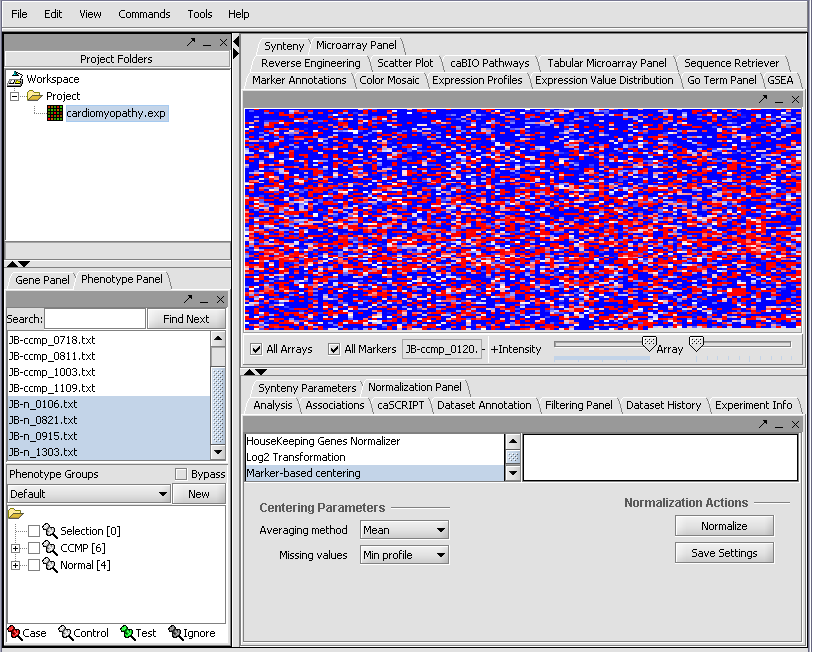
Resulting in the following colorful display of the array data for the first array....
Your File Nodes will now be Merged into one Project folder.