GeWorkbench Release 2.2
From Informatics
General notes on previous geWorkbench releases
- General notes, feature requests and FAQ page - This page was started with material from the time of release 1.7.0 and will be updated continually.
- Links to other release pages:
Other geWorkbench planning pages
- geWorkbench Development Notes - contains notes for several changes that were part of release 2.2.0.
- GeWorkbench TODO List - items that need to be planned or documented.
- Nikhil's page for geWorkbench web notes: http://wiki.c2b2.columbia.edu/informatics/index.php/GeWorkbench-Web
- The geWorkbench Roadmap (local version) contains possible directions for future development.
- caBIG has a separate geWorkbench Roadmap page that we must maintain.
- caBIG/NCI also provides the official download page for geWorkbench
Release Schedule for 2.2.2
- Actual release date: 8/19/2011
Release Schedule for 2.2.1
- geWorkbench 2.2.1 code freeze: 6/30/2011
- System testing started: 7/1/2011
- System Testing concluded: 7/7/2011
- Bug fixes concluded: 7/21/2011 (well not really!)
- Final release target: 7/28/2011
- Actual release date: 7/29/2011
Release Schedule for 2.2.0
- geWorkbench 2.2.0 code freeze: 4/22/2011
- System testing started: 4/25/2011
- System Testing concluded: 4/29/2011
- Final release target: 5/17/2011
- Actual release date: 5/24/2011
Role Assignments
- Release Manager – Kenneth Smith
- Release Engineer – Zheng Ma
- Tech Lead – Zhou Ji
- Tester – Udo Többen, and the rest of the bunch
- Test Manager – Udo Többen
- Technical Writer – Mary VanGinhoven
Things to remember
- Best practices for defect management - See also Aris's email of 8/20/09 on this topic.
- geWorkbench Roadmap page at NCICB - keep up to date with actual plans and developments - at https://cabig-kc.nci.nih.gov/Molecular/KC/index.php/GeWorkbench_Roadmap
- InstallAnywhere JRE update packs: http://www.flexerasoftware.com/products/installanywhere/files-utilities.htm
- Our local caArray is at afapp1.c2b2.columbia.edu port 38080 (web interface).
Updates to external components 2.2.2
- None.
Updates to external components 2.2.1
- GeneOntology OBO file- (6/29/2011) Updated to this date. But geWorkbench now downloads latest each time.
- JASPAR - (checked 6/29/2011) As of today, there has been no update to the JASPAR motif files since October 12, 2009. we use the following files from the JASPAR CORE SQL tables directory (http://jaspar.genereg.net/html/DOWNLOAD/jaspar_CORE/non_redundant/all_species/sql_tables/):
- MATRIX.txt
- MATRIX_ANNOTATION.txt
- MATRIX_DATA.txt
- JMol - (6/29/2011) Updated to JMol 12.0.45.
Updates to external components 2.2.0
- JMol - (3/23/2011) Updated to JMol 12.0.35. Note that 12.0.36 and 12.0.37 had a bug in their right-click menus, so we don't use them.
- GeneOntology OBO file- (4/19/2011) Updated to this date. But geWorkbench now downloads latest each time.
- GSEA - (3/23/2011) Updated our GenePattern server with latest GSEA module, needed for connectivity to Broad server.
- JASPAR - (3/23/2011) As of today, there has been no update to the JASPAR motif files since October 12, 2009.
geWorkbench 2.2.0 Grid Service URLs
The default Index Service and Dispatcher Service are hard-coded in configuration file "conf/application.properties". Updating these defaults is part of the release process. That is, for the production version, the production URLs must be entered.
Production URLs for 2.2.2
Index and Dispatcher
- Default Index Service: http://cagridnode.c2b2.columbia.edu:8080/v2.2.2/wsrf/services/DefaultIndexService
- Default Dispatcher Service: http://cagridnode.c2b2.columbia.edu:8080/v2.2.2/wsrf/services/cagrid/Dispatcher
Note - these "v2.2.2" URLs actually point to the same services as v2.2.1. There were no changes to grid services in 2.2.2.
Standard geWorkbench Grid Services
where ServiceName is e.g. Anova, Aracne, etc.
MarkUs and Skyline Service URLs
- bhapp.c2b2.columbia.edu:8080/wsrf/services/cagrid/MarkUs
- bhapp.c2b2.columbia.edu:8080/wsrf/services/cagrid/SkyLine
MarkUs and Skyline RESULT URLS
Both MarkUs and Skyline pick reference a web service to retrieve results. The results remain on the remote server and only the information requested is returned to geWorkbench. As a consequence, the results of these two analyses cannot be preserved indefinitely, even by saving a workspace.
The result urls are independent of cagrid index/dispatcher url, but linked tightly to bhapp.c2b2.columbia.edu.
Markus result url: http://bhapp.c2b2.columbia.edu/MarkUs/cgi-bin/browse.pl?pdb_id=MUS... [^] It's url for MarkUs web site, and won't change when we move our services around.
Skyline result url: http://cagridnode.c2b2.columbia.edu:8080/luna/SkyLineData/output [^] which is a proxy forward to bhapp.c2b2.columbia.edu:8080/SkyLineData/output
When cagrid index service moves to a new server, we just need to change the tomcat configuration of these two services to register to new index service. No need to change geworkbench code for them to work.
Production URLs for 2.2.1
Index and Dispatcher
- Default Index Service: http://cagridnode.c2b2.columbia.edu:8080/v2.2.1/wsrf/services/DefaultIndexService
- Default Dispatcher Service: http://cagridnode.c2b2.columbia.edu:8080/v2.2.1/wsrf/services/cagrid/Dispatcher
Standard geWorkbench Grid Services
where ServiceName is e.g. Anova, Aracne, etc.
MarkUs and Skyline Service URLs
- bhapp.c2b2.columbia.edu:8080/wsrf/services/cagrid/MarkUs
- bhapp.c2b2.columbia.edu:8080/wsrf/services/cagrid/SkyLine
Production URLs for 2.2.0
Index and Dispatcher
- Default Index Service: http://cagridnode.c2b2.columbia.edu:8080/v2.2.0/wsrf/services/DefaultIndexService
- Default Dispatcher Service: http://cagridnode.c2b2.columbia.edu:8080/v2.2.0/wsrf/services/cagrid/Dispatcher
Standard geWorkbench Grid Services
MarkUs and Skyline Service URLs
- luna.bioc.columbia.edu:8080/wsrf/services/cagrid/MarkUs
- luna.bioc.columbia.edu:8080/wsrf/services/cagrid/SkyLine
Production URLs from 2.1.0 and 2.0.0
These must be used for testing MarkUs and Skyline in the development version of geWorkbench 2.2.0.
- Default Index Service: http://cagridnode.c2b2.columbia.edu:8080/v2.0.0/wsrf/services/DefaultIndexService
- Default Dispatcher Service: http://cagridnode.c2b2.columbia.edu:8080/v2.0.0/wsrf/services/cagrid/Dispatcher
Development URLs
All grid services except MarkUs and Skyline use the development index service and dispatcher, and have development grid services.
- Default Index Service: http://afdev.c2b2.columbia.edu:8080/wsrf/services/DefaultIndexService
- Default Dispatcher Service: http://afdev.c2b2.columbia.edu:8080/wsrf/services/cagrid/Dispatcher
System Testing
See results at http://afdev.cgc.cpmc.columbia.edu/systemtest/BrowseLogs.php
Known caArray issue that keeps coming up
There is a problem with the caArray server code, in that as long as a geWorkbench session is running, the server retains the last used username/password, if any have been submitted.
See bugs 2022, 2555.
Two problems can arise:
Problem 1
Unfortunately, that defect still exists in the new API. The only situtation you will see it in is as follows:
- User A connects to caArray via geWorkbench and enters his/her credentials.
- An anonymous user then connects to caArray using the same geWorkbench instance. This anonymous user can still see User A's protected data.
The bug does not affect any other situations. E.g., if the users are using different instances of geWorkbench, there is no problem. If the second user is passing in a new set of credentials, it's not a problem. It is only a problem when the first user is credentialled and the second user is anonymous, and they are both connecting through the same geWorkbench one after another.
Thanks! Rashmi
Problem 2
Once a username and password have been entered and submitted to caArray, you cannot go back to using no username/password, except by restarting geWorkbench. However you can still put in a different username/password combination. This is a property of the caArray server-side code. Thus if you have no valid username/password and enter an incorrect one, you will need to restart geWorkbench before you can query caArray public experiments again (no login required).
Major changes in release 2.2.0
- CNKB - #2389 - export complete CNKB interactomes (SIF or ADJ formats).
- Cytoscape
- #2424 - 1) calculate the Pearson's correlation coefficient for the expression profiles of each pair of nodes connected by an edge in an interaction network. Filter edges to display based on the magnitude of the correlation coefficient.
- #2424 - 2) create a new subnetwork containing only edges exceeding correlation threshold calculated in (1).
- #2429 - From an existing network, create a subnetwork containing only nodes in a marker set defined in the Markers component.
- File Parsers - #2388 - import an ARACNe adjacency matrix from a file.
- Filters
- #2444 - Multiple probeset per gene filter - for genes with multiple probesets (markers), remove all but one probeset based on: retain only (a) highest coefficient of variation, (b) highest mean, (c) highest median.
- #2445 - Multiple Entrez GeneID Filter: Filter out markers which are annotated to (a) no Entrez gene id, or (b) multiple Entrez gene ids.
- Fold-change Analysis - #2431 - a new component that performs fold-change analysis and places markers that pass the specified threshold into two new sets in the Markers component: 1 for markers with positive fold-change, and the other for those with negative fold-change.
- Gene Ontology
- The Gene Ontology component is now always available when a microarray dataset has been loaded along with its annotation file. The GO Tree can be browsed or searched for any term. The markers annotated to any term can be returned to a new set in the Markers component.
- #1875 - most recent Gene Ontology OBO file now downloaded automatically from internet when geWorkbench started, with option to instead use a specified file from disk.
- GenePattern GSEA - analysis component added.
- Master Regulator Analyis - #2523 and others. Master Regulator Analysis component fully revised. Now allow any list of markers to be used for the phenotype signature. Bar code graph revised to match style of published work.
Other changes
- MEDUSA component's local version is restored. It was disabled when we switched to Java 6.
- Gene Ontology view for expression data
- caArray password is encrypted when cached.
- package geWorkbench core in seven jar files
- geWorkbench core's parser/format code has the package name changed, mainly dropping the 'components' level in the package path
- remove many not-used jar files (libraries)
- a series of MicroarrayViewEventBase/AnalysisPanel related code cleaning-ups
- general code clean-up (eliminate unnecessary event publish/subscription, project panel, context manager, position histogram, etc)
- ARACNe - #2366 - in ARACNe, bootstrapping is re-enabled but only single threaded.
- ARACNe - #2482 - ARACNe results can now be pruned to retain only highest MI edge per gene-gene pair, or return all edges.
- BLAST - #2419 - continued improvements to BLAST interface to match NCBI website functionality and to improve usability.
- CCM - Sequence Analysis -> BLAST Analysis, Alignment Viewer -> BLAST Alignment Viewer.
- Gene Ontology Viewer - #2391 - When a marker set is returned for a GO term, the set is given the term name.
- GEO Soft - #2402, #2462, #2465 - GEO Soft parsers improved to handle various special cases - multiple platforms, missing values, mixed sample and data matrix files...
- Grid Services - #1773 - simplified grid service activation (removed one radio button).
- JMOL - #2505 - updated to JMOL version 12.0.35.
- Markers/Arrays component - #2430 - dynamic filtering of displayed marker or array list as search term is entered.
- MarkUs
- #2500 - Add ability to retrieve prior MarkUs jobs by job id
- #2509 - add private key option to MarkUs job submission
- MINDy - #2214 - corrected sign of modulation effect in table displays.
- Pattern Discovery #2119 - corrected display problems in Pattern Discovery related to regular expressions and use of substitution matrices.
- Preferences - #2393 - Added ability to reorder data sorted by marker name, gene name, or original order (set in preferences, affects all components).
- Sequence Retriever - #2518 - fixed problem with obtaining name of latest human genome build from Santa Cruz.
- Tabular Microarray Viewer - #2253 - Tabular Microarray viewer now allows adjustable precision in display, and choice of fixed or scientific notation.
- t-test - #1626 - changed math package used to correct precision problem with p-value calculation at very small p-values.
- Volcano Plot - #2492 - extreme point color range corrected.
Changes to documentation
- MINDy - In the tutorial and Online Help, the description of Table->Discretized Scores->Score value incorrectly described a range of -1 to +1 for the raw delta MI scores. The actual bounds on MI are zero to +infinity, so this phrase was removed in both sources.
- File Formats - Bug 1624 - "duplicate entries in annotation file should be reported" - added new section to "File Formats" page showing screenshot of error dialog offering 3 choices of action.
- Local Data Files - updated descriptions of GEO files.
- CNKB - all screenshots on tutorial page updated to reflect
- Don't use multiple markers from same gene because they all get the same query results, and the interaction totals reflects all of the duplicates.
- New default color scheme of Cytoscape 2.7
- New right-click menu options in Cytoscape 2.7.
- Cytoscape - Added new section to show all new features for release 2.2 - Correlation Overlay, t-test overlay, subnetworks etc.
- Data Subsets - Arrays
- Many new and revised screenshots added.
- Added new full section on visual properties editor.
- Now call the collections of sets "Lists" (pulldown menu entries).
- Much material was previously in a separate "Examples" section; this has now all been moved into the primary description of each menu item.
- Data Subsets - Markers
- All the same changes made to "Data Subsets - Arrays" were also made to "Data Subsets - Markers".
- Material unique to the Markers component was added as needed.
- Menu Bar - New tutorial written to cover all actions available in the top level menu bar.
- ARACNe
- Corrected and improved descriptions of DPI Tolerance and DPI Target List.
- Added Technical Notes section and usage notes.
- Mapped relationship of ARACNe command line options -s and -l to ARACNe in geWorkbench.
- Rewrote introduction.
- SVM - all new chapter written. It is updated to reflect small changes to the Test tab GUI in development.
- Pattern Discovery - all screenshots replaced. Changes reflect:
- Fixed display of regular expression matches in Full Sequence view after running with "Exact" unchecked.
- In Advanced tab, matrix and threshold settings now disabled when "Exact" is checked.
- Old screenshots misspelled "Exhaust.".
- Better illustration of multiple pattern displays in viewer.
- Once most screenshots were replaced for above reasons, rest were replaced because could see the difference in new vs old "Look and Feel".
- Local Data files - full update, including
- updated screenshots and descriptions,
- added additional material about file browser, merging, annotation files....
- Projects
- Improved descriptions and added additional screenshots throughout
- Added missing options such as RCSB PDB.
- Add new section on Workspaces
- t-test
- Fully revised all sections.
- Added sections on Volcano Plot and Color Mosaic
Major changes in release 2.2.1
- ANOVA
- Control buttons repositioned for easier use.
- ARACNe
- (#2648) - The adjacency matrix is no longer made symmetric. Each line now starts with a hub gene.
- Similarily, the adjacency matrix, when saved to file, is not made symmetric.
- Bonferroni correction option added.
- Merging of multiple probesets is now just within a hub gene, not global.
- Color Mosaic
- Export button and right-click menu - both now support export of significance results (p-values) from both t-test and ANOVA.
- CNKB
- Export of network adjacency matrix from Project Folder omitted gene/probeset names.
- Cytoscape
- (#2589, #2638) - If a network might be too large to view in Cytoscape, the user is offered an alternate visualization (text). The threshold for warning can be changed by the user. This replaces warnings that were previously built in to several components e.g. ARACNe. Now, an adjacency matrix is always created, regardless of size.
- (#2597, #2598, #2599) - some inconsistencies in how selected nodes in Cytoscape were reflected as markers in the Markers component were found and fixed.
- t-test overlay on network. This did not work in release 2.2.0 due to a late change. Fixed.
- Fold-Change Analysis - New component accidentally omitted from release 2.2.0.
- Network Import
- Networks can now be imported either from Adjacency matrix files (#2432) or from SIF-format (#2434) files. The networks can be represented by gene symbols, Entrez IDs, or probeset names, or by some other identifier (#2432). Networks are represented in memory with the same identifiers that were imported. That is, if a network is read in using gene symbols, then it is represented at this level within geWorkbench. Probesets for each gene can be obtained as needed for particular analyses.
- On import, .SIF and .ADJ file extensions are automatically recognized.
- On export, .ADJ is automatically added.
- Gene Ontology Analysis
- (#2512) - The analysis now supports using an alternate annotation file rather than an Affymetrix annotation file. Organism annotation files from the geneontology.org website can now be used.
- MarkUs
- (#2622) - Added support for job description and email notification fields.
- Master Regulator Analysis
- (#2610) - MRA can now be run from gene-level adjacency matrices created by CNKB.
- (#2301) - When saving results (target lists for MRs), a ".csv" file chooser is used and will append this extension automatically.
- MINDy
- (#2314) - An inconsistency in how modulators and targets were selected/deselected with respect to the "Displayed Targets Filter" was fixed.
- Pattern Discovery
- Problems that occurred when a run was canceled or the server unreachable were fixed.
- Project Folders
- (#2626) - Mousing-over a network node will display the number of nodes and edges in hover text.
- (#2646) - When saving a network, the ".adj" file filter will append ".adj" to the file created.
- Sequence Retriever (#2630, #2631, #2632)
- For DNA, now queries the "refGene" table rather than UCSC "known genes", so can now retrieve genomic sequence for all UCSC supported species.
- The transcription start point was off by one in query and display.
- Identifiers for retrieved sequences were being truncated if long.
- The "local" option for sequence retrieval was removed.
- The sequence location "spinner" controls can now be set to zero, and step in increments of 100.
- geWorkbench Installer - When run in Windows, the installer version of geWorkbench now displays its correct icon in the Windows task bar.
Components
This list copied directly from release 2.1.0.
List of Included Components
Noted on SVN refresh on 8/11/2010 - a number of outdated components look like dropped from SVN. See 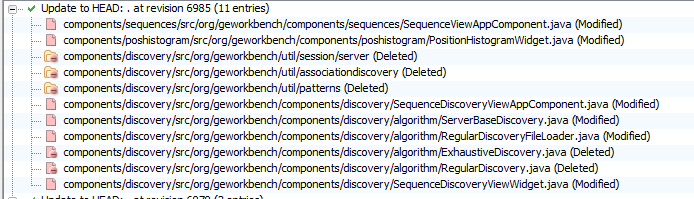
Data Managmenent:
- Arrays/Phenotypes
- Markers
- Preferences
- Project Panel
- Session manager - no one knows what this is - probably a SOAP interface. But it is definitely needed!
File input formats
- Affy File Format
- CEL File Loader
- Exp. Format
- FASTA Format
- Genepix File Format
- PDB Structure Format
- Tab-delimited (RMA Express Format)
Connectivity
- caArray2 - updated to support caArray 2.3.0 in release 1.8.0 (released September 2009). The caArray client jar is NOT backwards-compatible with any previous versions.
Data filters
- Filtering
- Affy Detection Call Filter
- Coefficient of Variation (new)
- Deviation Filter
- Expression Threshold Filter
- Genepix Filter (Two channel filter)
- Genepix Flag Filter
- Missing Values Filter
- Multiple probeset per gene filter
- Multiple Entrez GeneID Filter
Normalization
- HouseKeeping Genes Normalizer
- Normalization
- Log2 Tranformation
- Marker Centering Normalizer
- Mean Variance Normalizer
- Missing Values (Normalizer)
- Microarray Centering Normalizer
- Quantile Normalizer
- Threshold Normalizer
Experiment Information
- Dataset Annotation
- Dataset History
- Experiment Info
- Version Information
Analyis/Visualization
- Alignment Results
- Analysis
- ANOVA
- ARACNe2 - adds Adaptive Partitioning algorithm and Preprocessing mode.
- caBIO Pathways (this has been integrated in the Marker Annotations component)
- Cancer Gene Index integration in the Marker Annotations component.
- CELImageViewer
- Cellular Networks Knowledge Base
- Color Mosaic
- Component Configuration Manager.
- Cytoscape_V2_4 - updated version of Cytoscape.
- Dendrogram
- Expression Profiles
- Expression Value Distribution
- Fold-change Analysis - by accident omitted from release 2.2.0. To be released in 2.2.1.
- Gene Ontology Enrichment Analysis and Display
- Hierarchical Clustering Analysis
- genSpace collaborative framework
- Image Viewer
- Jmol
- Marker Annotations
- MarkUs - Analysis and Viewer
- MRA - Master Regulator Analysis
- MatrixREDUCE
- Microarray Viewer
- MINDy - Analysis and Viewer
- Pattern Discovery
- Position Histogram
- Pudge
- Promoter
- Scatter Plot
- Sequence
- Sequence Alignment
- Sequence Retriever
- SOM Analysis
- SOM Clusters
- t Test Analysis
- Tabular Microarray Viewer
- Volcano Plot
- GenePattern components
- PCA (GenePattern) - Analysis and Viewer
- K-nearest neighbors (GenePattern)
- SVM 3.0 (GenePattern) - Analysis and Viewer - include, we need to develop online help and tutorial (Aris).
- WV - Weighted Voting (GenePattern)
- GSEA
- GeneWays (need to update status for release 2.0) (need for Cytoscape viewer for ARACNe)- this component not working; we do
not have end-user documentation materials (Aris). However, Geneways must be included for Cytoscape interaction to function, but Geneways itself cannot be chosen as a component nor used directly.
Excluded and Dropped Components
The release creation script in build.xml now explicitly includes components by name (previously it excluded components by name) The following is a list of modules known to be excluded.
Excluded components
The following components are excluded for a variety of reasons, most often due to lack of formal requirements documentation or/and associated system test scripts. Some of them should be scheduled for inclusion in the next production release. For modules not found in the current all.xml a path to the component is shown.
Still under development:
- CART (GenePattern) - this component has not yet been released. Is part of another component and must be excluded manually from the final installer release build.
- Cancer-GEMS (awaiting further development from NCI)
- NetBoost
- EdgeListFileFormat (NetBoost)
- Evidence Integration
- IDEA - finished development but not yet tested for release.
- MEDUSA
Not actively being developed:
- GCRMA Via R CEL Loader (in \geworkbench\src\org\geworkbench\components\parsers)
- Multi-t-test (OK, but need to understand when it would be used, e.g. after ANOVA, and if it is what we really want).
- SMLR - Sparse Multinomial Logistic Regression - implementation by John Watkinson.
- SVM Format (in \geworkbench\src\org\geworkbench\components\parsers) (left over from a John Watkinson project).
- Synteny (in \geworkbench\components\alignment\src\org\geworkbench\components\alignment\client)
- t-profiler
- caScript
Dropped components
These components are not expected to be used again.
- CuteNet (GeneWays)
- Column Major Format (in \geworkbench\src\org\geworkbench\components\parsers)
- Frequency Threshold Filter (There is a class called AllelicFrequencyThresholdFilter in \geworkbench\components\filtering\src\org\geworkbench\components\filtering)
- GeneOntology (the original component, now replaced by geneontology2/Ontologizer2.0)
- Genotypic File Format (in \geworkbench\src\org\geworkbench\components\parsers\genotype)
- Network Browser (was part of Reverse Engineering - would require major rewrite to revive. PathwayDecoder is module name)
- Pattern Discovery Algorithm (association analysis)
- Patterns (Pattern Panel) - Omit from release - Appears to have been superseded by the Sequence component.
- Reverse Engineering (non-ARACNE, unpublished algorithm. PathwayDecoder is module name)
- Simulation (a student project)
Note - the original "interactions" component was dropped and reimplemented as the Cellular Networks Knowledge Base. It took a brief detour as being called component "interactions2".
Externally supplied components
The following components originate external to the geWorkbench source tree:
MatrixReduce
Source
MatrixReduce source code was obtained from the Bussemaker lab and a modified copy saved under: adcvs.cu-genome.org:/cvs/magnet/matrixreduce_distribution. This modified copy contains Java API changes made to integrate with geWorkbench.
Compiling
MatrixReduce is compiled using the following commands:
- FitModel binary is compiled manually as follows
- gcc -c -O2 -mno-cygwin -funroll-loops *.c
- gcc -mno-cygwin -static nrutil.o fncs_cmns.o fncs_seqs.o fncs_tdat.o fncs_seed.o fncs_app1.o fncs_app2.o fncs_nrcs.o fncs_topo.o fncs_mylm.o fncs_bits.o FitModel.o -o FitModel –lm (for windows and linux)
- gcc -mno-cygwin nrutil.o fncs_cmns.o fncs_seqs.o fncs_tdat.o fncs_seed.o fncs_app1.o fncs_app2.o fncs_nrcs.o fncs_topo.o fncs_mylm.o fncs_bits.o FitModel.o -o FitModelMac –lm (for Mac)
- API jar: The Java API jar is created with the makefile, command "make jar".
- FitModel binary is compiled manually with gcc, with extra flags to tell it to not use Cygwin, to optimize and to unroll loops
- FitModel.exe bundles both the NR (Numerical Recipies) and GNU libraries.
- The API jar is created with the makefile under MatrixREDUCE's top directory.
Notes
See comment on white spaces in file names/paths in Mantis : http://mantis.cu-genome.org/view.php?id=1316
Aracne.jar for MINDY
Although ARACNE is a geWorkbench component, the MINDY component uses a version of ARACNE that is externally maintained. The file aracne.jar is copied directly into the geWorkbench CVS tree.
The location of the external ARACNE code is:
The version of the external ARACNE code is:
MINDy jar file for caGrid
- Source tree is kept in the geWorkbench local CVS repository.
- Current version is MINDY-0.3.jar
- Compile with ant dist-jar. The final jar file will be in the "dist" directory.
- Compile using Java 1.5 for compatibility with caGrid 1.3.
- If we ever change to caGrid 1.4, the we can compile MINDy with Java 1.6.
Any other components?
Analysis components - external runtime dependencies
These analyses depend on C2B2 services,either via grid or servlets. External services like BLAST are not included here.
| component | local | external type | username/password | known to work outside campus |
|---|---|---|---|---|
| ANOVA | yes | grid | grid_default | ? |
| ARACNe | yes | grid | grid_default | ? |
| CNKB | no | servlet | some open, some protected data | ? |
| GenSpace | no | servlet | genSpace account | ? |
| GSEA | no | GenePattern | instance-specific | prob not, use Broad etc. |
| Hierarchical Clustering | yes | grid | grid_default | ? |
| KNN | no | GenePattern | instance-specific | ? |
| MarkUs | no | servlet, grid | open | ? |
| MINDy | yes | grid | grid_default | ? |
| MRA | yes | no | - | not applicable |
| MatrixREDUCE | yes | grid | grid_default | ? |
| PCA | no | GenePattern | instance-specific | ? |
| PUDGE | no | servlet | open | ? |
| SkyBase | no | grid | open | ? |
| SkyLine | no | grid | grid_default | ? |
| SOM | yes | grid | grid_default | ? |
| SVM | no | GenePattern | instance-specific | ? |
| WV | no | GenePattern | instance-specific | ? |