Previous Release Issues
Contents
- 1 geWorkbench 2.5.1
- 2 geWorkbench 2.5.0
- 3 geWorkbench 2.4.1
- 4 geWorkbench 2.4.0
- 5 geWorkbench 2.3.0
- 6 geWorkbench 2.2.2
- 7 geWorkbench 2.2.1
- 8 geWorkbench 2.2.0
- 9 geWorkbench 2.2.0 and previous
- 10 geWorkbench 2.1.0
- 11 geWorkbench 2.1.0 and previous
- 12 geWorkbench 2.0.2
- 13 geWorkbench 2.0.1
- 14 geWorkbench 2.0.0
- 15 geWorkbench 1.8.0 and earlier
- 16 geWorkbench 1.7.0
geWorkbench 2.5.1
R-based analyses (SAM and VIPER) on Windows platforms fail if space in path
The Problem
If you are using a version of geWorkbench on Windows which does not include the built-in JRE, then you may experience problems with components that make use of R. This problem began with a change in Java 6 update 45, and also was introduced in Java 7 update 21. The problem lies in a change in how spaces in file paths are handled, e.g. "C:\Program Files\R".
Note - the problem has been resolved in the geWorkbench 2.5.2 branch.
Background
geWorkbench for Windows can be downloaded with or without a built-in JRE. The version of geWorkbench with built-in JRE does not experience the problem, because it uses Java 6 update 26.
However, you may wish to use a version of geWorkbench which does not include the built-in JRE, for example in order to use a 64-bit version of Java.
Workaround
Install R into a directory path that does not have any spaces in it, e.g. C:\R\R-3.0.3. You can then run geWorkbench with the most recent versions of Java.
MRA-FET
Running MRA-FET with an activated marker set will give incorrect results. Please do not activate any marker sets when running MRA-FET (Mantis issue #2793). Fixed in 2.6.0.
VIPER
A file needed by VIPER was omitted in geWorkbench 2.5.0 and the initial release of geWorkbench 2.5.1. A new version of 2.5.1 containing the missing file was made available on November 12, 2013 at 8 pm EST on the download server. There were no source code changes.
MarkUs
A problem with the MarkUs service URL has been resolved through a redirect to the new URL, https://bhapp.c2b2.columbia.edu.
Update - this redirect does not seem to be working anymore. To see your results, enter the MarkUs job into a web browser as follows, using the job name returned for the MarkUs results node in the Workspace:
https://bhapp.c2b2.columbia.edu/MarkUs/cgi-bin/browse.pl?pdb_id=MUS#### where #### represents the particular job number returned in geWorkbench for your job.
These problems are fixed in the upcoming 2.6.0 release of geWorkbench.
Windows Control Panel Version Number
The initial release of geWorkbench 2.5.1 reported its version as 2.5.0 in the Windows control panel. This was corrected in the new copy of 2.5.1 made available November 12, 2013 at 8 pm on the download server.
geWorkbench 2.5.0
- Workspace (#3577) - (fixed in 2.5.1) The "merge" function for expression datasets is not working when invoked from the "File" menu. However, it does work if two or more expression datafiles of the same type are loaded together in the same file open operation. To do this, the files should be in the same directory, and all files to be merged should be selected together (multiple-selection). The files will then be merged into a single dataset and placed into the Workspace.
geWorkbench 2.4.1
BLAST - Due to a change in the output from the NCBI BLAST server, sequence alignments are not being displayed. This will be fixed in version 2.5.0 by switching to using the XML results file.
Marker Annotations - As of September 30th, 2013, NCI-CBIIT is expected to deactivate the caBIO service on which the geWorkbench Marker Annotation component depends. geWorkbench 2.5.0 uses the bioDBNet service as its data source instead.
geWorkbench 2.4.0
The following problems have been noted in geWorkbench 2.4.0. Some may apply to earlier releases as well.
- BLAST (9/26/2012) - due to a change in the HTML result format from the NCBI website, BLAST in geWorkbench 2.4.0 and all previous versions no longer works.
- ARACNe2 -
- (#2885) ARACNe and MINDy, if local job cancellation is requested, call a non-safe method, thread.stop(), which can occasionally produce an exception.
- Java 7 - geWorkbench 2.4.0 was developed and tested using the Java 6 JDK and JRE. Although several incompatibilites with Java 7 were identified and fixed, as least one remains, shown below. For this reason, please only use Java 6 JREs when running geWorkbench 2.4.0 or earlier.
- Marker Annotations - (#3072) does not receive activated marker set, caBIO client library conflict with Java 7.
- BLAST - (#2825) the maximum number of hits returned is 50, even if more were requested.
- caArray - (#2893) due to a synchronization problem, a large empty white space may occasionally appear in the caArray list of experiments.
- Cytoscape
- (#2843) if an instance of Cytoscape is running on a machine, and then Cytoscape is also loaded in geWorkbench, you may see a warning "org.cytoscape.coreplugin.cpath2.http.HTTPServer[WARN]: HTTPServer couldn't create socket.".
- (#2716) Direct LinkOuts from the right-click menu to UniProt may not work. However, they do work when selected from the Cytoscape Data Panel.
- GenePattern connectivity (#2701) - if you successfully connect to a GenePattern server, and later that server is no longer available, you will experience a time-out delay. This was seen when first connecting to a GP server through a firewall via VPN, then trying again without the VPN.
- GenomeSpace - transfer of large data files is not supported. This has been fixed in development (Mantis issue #3273).
- Grid Services/Dispatcher Service:
As of release 2.3.0, outbound data (from geWorkbench to the grid service) is moved using caTransfer, both from geWorkbench to the Dispatcher, and from the Dispatcher to the grid service. Returning data is transferred for both hops as a base-64 encoded string, which for very large data sets can lead to an out-of-memory error. Full replacement with caTransfer is being investigated.
The error message will read:
Out-of-memory error: Java heap space It is advisable to restart geWorkbench. You may also wish to increase the geWorkbench memory size.
In this case, as the message states, it is advisable to restart geWorkbench. Instructions for increasing the amount of memory available to Java are available in the geWorkbench FAQ. For Java memory requests larger than about 1.5 GB, running geWorkbench using a 64-bit Java Runtime Environment (JRE) will be necessary.
geWorkbench 2.3.0
The following problems have been noted in geWorkbench 2.3.0. Some may apply to earlier releases as well.
- ARACNe2 -
- (#3039, #3040) In all versions of geWorkbench up to 2.3.0 there is a problem with the implementation of the bootstrapping mode for ARACNe2. Bootstrapping should not be used. This will be fixed in release 2.4.0. The sample with replacement step was discarding duplicates, resulting in a reduced data set with only unique arrays.
- (#2885) ARACNe and MINDy, if local job cancellation is requested, call a non-safe method, thread.stop(), which can occasionally produce an exception.
- Java 7 - geWorkbench 2.3.0 was developed and tested using the Java 6 JDK and JRE. Subsequent testing with Java 7 has shown a number of problems, shown below. For this reason, please only use Java 6 JREs when running geWorkbench 2.3.0 or earlier.
- CNKB - (#3008) activated markers not transfered from Markers component (fixed in development).
- Color Mosaic - (#3011) activated array set causes color mosaic display to turn red (fixed in development).
- Expression profiles - (#2980) activated array set causes expression profile not to be drawn (fixed in development).
- Microarray Viewer - (#3071) no display if marker set activated (fixed in development).
- Marker Annotations - (#3072) does not receive activated marker set, caBIO client library conflict with Java 7.
- IDEA - (#3001) Some edges appear twice in the results list, differing only in node order: N1-N2 and N2-N1.
- BLAST - (#2825) the maximum number of hits returned is 50, even if more were requested.
- caArray - (#2893) due to a synchronization problem, a large empty white space may occasionally appear in the caArray list of experiments.
- Cytoscape
- (#2843) if an instance of Cytoscape is running on a machine, and then Cytoscape is also loaded in geWorkbench, you may see a warning "org.cytoscape.coreplugin.cpath2.http.HTTPServer[WARN]: HTTPServer couldn't create socket.".
- (#2716) Direct LinkOuts from the right-click menu to UniProt may not work. However, they do work when selected from the Cytoscape Data Panel.
- GenePattern connectivity (#2701) - if you successfully connect to a GenePattern server, and later that server is no longer available, you will experience a time-out delay. This was seen when first connecting to a GP server through a firewall via VPN, then trying again without the VPN.
- Grid Services/Dispatcher Service:
As of release 2.3.0, outbound data (from geWorkbench to the grid service) is moved using caTransfer, both from geWorkbench to the Dispatcher, and from the Dispatcher to the grid service. Returning data is transferred for both hops as a base-64 encoded string, which for very large data sets can lead to an out-of-memory error. Full replacement with caTransfer is being investigated.
The error message will read:
Out-of-memory error: Java heap space It is advisable to restart geWorkbench. You may also wish to increase the geWorkbench memory size.
In this case, as the message states, it is advisable to restart geWorkbench. Instructions for increasing the amount of memory available to Java are available in the geWorkbench FAQ. For Java memory requests larger than about 1.5 GB, running geWorkbench using a 64-bit Java Runtime Environment (JRE) will be necessary.
- MINDy - (#2698) if the MINDy Heatmap window is resized, and then scrolled down/up, the heatmap does not redraw correctly. A redraw can be forced by switching between two different modulators. In earlier versions, a "Refresh" button was available to force the redraw.
- Duplicate Markers - microarray datasets should not contain duplicate marker names. If they do, duplicates will be silently removed. In a subsequent release, a warning will be issued.
- Macintosh - List multi-select: List multi-select does not work in geWorkbench 2.3.0 and earlier on the Mac. However, in geWorkbench 2.4.0 development version, list multi-select can be accomplished with command-click.
geWorkbench 2.2.2
- (#2830) - a parsing problem in tblastx results due to changes in the HTML returned by NCBI is fixed in 2.3.0. The number from column "N" was appearing after the score in the e-value column.
- (#2729) - all versions prior to release 2.3.0 were limited in the number of arrays that could be downloaded in caArray, usually less than 100, due to a problem with memory management. With release 2.3.0, there are now no issues with downloading large numbers of arrays. Downloads of more than 500 arrays have been tested without problem in release 2.3.0.
- (#2925) - In 2.2.2 and previous versions, if two experiments had the same name in caArray, data from only one could be downloaded. Fixed in 2.3.0.
geWorkbench 2.2.1
- Gene Ontology Analysis - if a workspace containing a microarray dataset and its annotation information is saved and then restored, the gene ontology analysis will not work properly and should not be used.
- Gene Ontology Viewer - In some cases when browsing the GO tree, when you select a particular term and view its associated genes, the list contains the "no gene name" entry (---). If this list is in turn copied to a marker set, the "---" will cause all matching markers, that is, those with no gene names, to be included in the new set.
- Workaround - use the "Multiple Entrez ID filter" to remove all markers that have no gene name associated with them.
- Installation - Windows 7 and Vista users are advised during installation to install to the "C:\geworkbench_2.2.1" directory. However, only users with administrator privileges can install to the c:\ root directory. Instead, users with regular user permissions should install to their own home directory, C:\Users\username\geworkbench_2.2.1, where username is your own login name.
geWorkbench 2.2.0
Cytoscape plugin
- t-test result overlay on network - A new feature in geWorkbench release 2.2.0 was the ability to superimpose the results of a t-test onto a network displayed in Cytoscape. Due to last minute changes before the release, this feature is not working in geWorkbench 2.2.0.
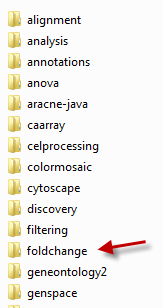
Fold Change Component
The Fold Change analysis component, which was a new component in release 2.2.0, was inadvertently omitted from the release. The component is available in this zip file: Foldchange.zip.
- Download the Foldchange.zip file.
- Find your geWorkbench 2.2.0 installation directory, and copy the zip file into the components directory. For example, on Windows 7, the components directory is likely located at "C:\geWorkbench_2.2.0\components".
- Unzip the file into the components directory, so that the "foldchange" directory appears in the components directory. It should look like this:
- The next time you start geWorkbench, the Fold Change component will be available.
PCA 3D plot
When using a 64-bit JVM, the Java 3D library requires OpenGL version 1.2 or higher to be supported by your display driver.
geWorkbench 2.2.0 and previous
- GO Terms Analysis - Only an Affymetrix-format annotation file can be used in the alternate annotation file field. This will be fixed to allow annotation files from the GO website to be used.
- caArray - Once a username and password have been entered and submitted to caArray, you cannot go back to using no username/password, except by restarting geWorkbench. However you can still put in a different username/password combination. This is a property of the caArray server-side code. Thus if you have no valid username/password and enter an incorrect one, you will need to restart geWorkbench before you can query caArray public experiments again (no login required).
- BLAST - The NCBI BLAST server may occasionally return an error when sequences are searched from geWorkbench. The problem appears to depend on the load on the NCBI BLAST server.
- GSEA (bug #2506) - On Windows (tested on Windows 7), using the 32-bit JVM, the following error message may appear in the error log. It can be disregarded.
- *** Error: nspr4.dll doesn't exist under C:\repository\svn\geworkbench\components\gpmodule_v3_0\lib\ielib
- 64-bit JVM known restrictions for GSEA, MarkUs, and Pudge (bug #2506 and others). A 32-bit JDIC browser (C++) interface (Java wrapper) is used. As currently compiled, the browser will not work with most 64-bit JVMs. The display will work correctly using a 32-bit JVM, such as included with the installer versions of geWorkbench.
- Windows - GSEA, Pudge and MarkUs will not display.
- Mac - prior to version 10.6: GSEA, Pudge and MarkUs will not display.
- Mac - version 10.6+, works for GSEA and Pudge. Does not work for MarkUs
- Linux - should work if Mozilla browser is installed.
- Solution - If you encounter this problem, switch to using a 32-bit JVM (included with all installer versions of geWorkbench except "Generic").
- A warning message will be displayed if this problem is encountered. It will print your operating system and architecture (e.g. amd64 for 64-bit systems), and provide a link by which to access the remote content directly in your web browser.
geWorkbench 2.1.0
- Sequence Retriever - (April 2011) The hg19 human genome build is not appearing in the list of genomes for DNA sequence retrieval. Being fixed in version 2.2.0.
- GEO Soft format files - two problems have been found in loading specific GEO Soft format data files.
- geWorkbench can only analyze data from one platform at a time. Some GEO Series (GSE) files can have data from multiple platforms, and opening one of these files this lead to the data being incorrectly read. A feature to select a single platform within such a file has been added in version 2.2.0. (Note that often you can find associated with such an experiment a "Series matrix format" file, which is platform specific.
- The geWorkbench GEO Soft format parser rejects a file with missing values. Fixed in version 2.2.0.
- SkyBase component could not retrieve data - RESOLVED - changes in December 2010 to the SkyBase backend database temporarily disabled the ability of geWorkbench to query it. This issue was resolved with changes to the backend database in January 2011.
- Linux installation error - A problem was seen when trying to install geWorkbench on Ubuntu Linux in a desktop configuration using the installer with included JRE. The same installer does work on another Linux variant. We are investigating. You may be able to use the Linux installer with no JRE instead - just make sure you have a current Java JRE installed on your system. See the geWorkbench installation instructions.
- t-test - the t-test incorrect reports permutation settings into the Dataset History when the t-distribution is used (no permutations involved). This is just a reporting issue. Fixed in version 2.2.0.
- Color Mosaic - The search by gene name or accession number (probeset id) is not working. Problems are seen both when viewing a t-test result and when viewing the main microarray dataset node. Fixed in version 2.2.0.
geWorkbench 2.1.0 and previous
- MINDy - starting with version 1.8.0, the sign of the modulator score shown in the Modulator, Table and List views was reversed. Fixed in 2.2.0.
geWorkbench 2.0.2
- Alignment Results (BLAST Results Viewer) - Due to recent changes in the NCBI BLAST website (August, 2010), geWorkbench could not retrieve hit sequences to add them to the project. This affected two buttons: "Add Selected Sequences to Project" and "Only Add Aligned Parts". Fixed in release 2.1.0
- Dataset History: the settings used by the following normalizers are not recorded in the Dataset History component: Mean Variance Normalizer, Missing Values Normalizer, Quantile Normalizer, Threshold Normalizer. Fixed in 2.1.0.
- CNKB - not all listed interaction databases are open for querying. See the page CNKB Data for a summary of available databases and interaction types. Fixed in 2.1.0.
geWorkbench 2.0.1
- genSpace - Logging was not working. Fixed in release 2.0.2.
geWorkbench 2.0.0
- Grid Services - No geWorkbench grid services are currently available outside of the Columbia firewall. Fixed in release 2.0.1.
- genSpace - Logging was not working. Fixed in release 2.0.2.
geWorkbench 1.8.0 and earlier
Linux on the desktop
Description of the problem
A problem has been found on the Linux desktop platform (as opposed to a remote Linux server) when running InstallAnywhere-packaged versions of geWorkbench with include the Java Run-Time Environment (JRE) (geWorkbench_v1.8.0_Linux_installer_with_JRE1.5.bin).
The conflict appears to involve the GTK toolkit used to draw components in the geWorkbench graphical user interface. This conflict apparently occurs with earlier versions of both the Java 5 JRE (JRE 1.5) and earlier versions of the Java 6 JRE (JRE 1.6).
The problem does not affect installations where geWorkbench runs on a remote Linux server and a Windows machine running a Cygwin X-windows server is used to view geWorkbench.
Resolution
We provide two versions of geWorkbench 1.8.0 without the packaged JRE. Either should work on Linux desktops if you separately install the latest version of the Java JRE. While formal testing of geWorkbench was performed using the Java 5 JRE (JRE 1.5), geWorkbench has also been shown to work with the latest Java 6 JREs (JRE 1.6).
The two versions for desktop Linux are:
- geWorkbench_v1.8.0_Linux_installer_noJRE.bin - This version includes an installation wizard but no JRE. You must install the latest JRE separately if it is not already on your system.
- geWorkbench_v1.8.0_Generic.zip - This version includes no installer wizard and no JRE. You just unzip the file with the "unzip" command. You must install the latest JRE separately if it is not already on your system.
Macintosh OSX and Java 6 JRE
Description of the problem
The Macintosh installer version of geWorkbench works correctly when Java 5 is used, but does not start when Java 6 is used.
Resolution
The Generic version of geWorkbench has been shown to work on Macintosh OSX 10.5 and the Java 6 JRE.
Windows and caArray downloads
Description of the problem
If a Java 6 JRE is used on Windows, e.g. with the Generic installation of geWorkbench, caArray downloads can lead to crashes.
Resolution
Use the Java 5 JRE, such as included with the Installer version of geWorkbench for Windows.
geWorkbench 1.7.0
- Annotations - The retrieval of annotations using the Marker Annotations component does not work for microarray data downloaded from caArray. Bug 1956.
- There is an easy work-around. After downloading data from caArray, and optionally setting up any needed groups of arrays or markers (e.g. treatment groups, controls etc), save the dataset as file type "Exp" format. You can then reload that saved dataset and associate the annotation file at that time.
- caArray - Data downloaded from caArray is being stored in a way that uses a large amount of memory, limiting the size of the dataset that can be used. Bug 1930.
- The first work-around is to increase the amount of memory allocated to geWorkbench.
- A second work around, if the first is impractical, is to download the data, using the "merge" check-box, in reasonable sized sets of arrays and then save each merged set as a separate "Exp" format file. Once saved, delete the downloaded data set in geWorkbench and then download the next set from caArray until all data has been saved to separate merged "Exp" files.
- The Exp files can then be all read back in together and merged into a single data object (make sure to check the "merge" check-box).
- caArray - Data downloads from instances of caArray housed at the NCI can be very slow.