Difference between revisions of "SVM"
| Line 1: | Line 1: | ||
| + | |||
| + | ==Introduction== | ||
The Support Vector Machines (SVM) module implements the support vector machines algorithm. It is a supervised classification method that computes a maximal separating hyperplane between the expression vectors of different classes or phenotypes. Given microarray data with ''n'' markers per sample, SVM outputs a hyperplane,''W'', which can be thought of as a vector with ''n'' components each corresponding to the expression of a particular marker. Loosely speaking, assuming that the expression values of each marker have similar ranges, the absolute magnitude of each element in ''W'' determines its importance in classifying a sample. | The Support Vector Machines (SVM) module implements the support vector machines algorithm. It is a supervised classification method that computes a maximal separating hyperplane between the expression vectors of different classes or phenotypes. Given microarray data with ''n'' markers per sample, SVM outputs a hyperplane,''W'', which can be thought of as a vector with ''n'' components each corresponding to the expression of a particular marker. Loosely speaking, assuming that the expression values of each marker have similar ranges, the absolute magnitude of each element in ''W'' determines its importance in classifying a sample. | ||
| + | |||
| + | |||
| + | ==Parameters== | ||
| + | |||
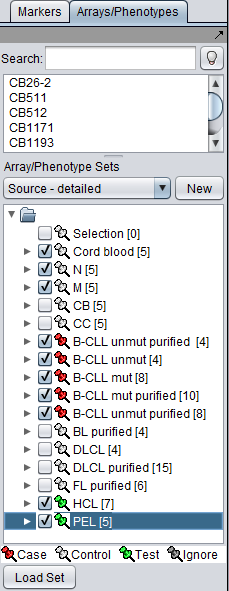
| + | The SVM module has no settable parameters for the computation. | ||
[[Image:SVM_Parameters_tab.png]] | [[Image:SVM_Parameters_tab.png]] | ||
| + | |||
| + | ==GenePattern Server Settings== | ||
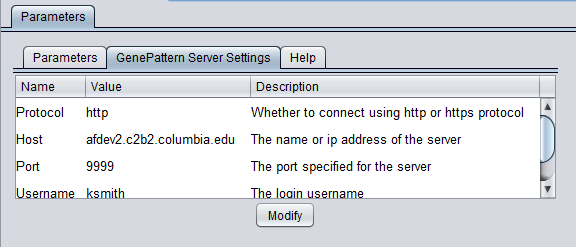
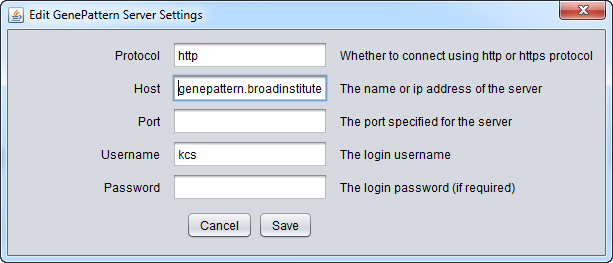
| + | To run GenePattern components, a GenePattern account is required. | ||
| + | |||
| + | * Protocol: HTTP or HTTPS, depending on the server being used. | ||
| + | * Host: URL of a GenePattern server. | ||
| + | * Port: Port at which the GenePattern server is located on the Host machine. | ||
| + | * Username: A valid user name on the specified GenePattern server. | ||
| + | * Password: A password, if required by the specified server. | ||
Revision as of 18:43, 7 March 2011
Introduction
The Support Vector Machines (SVM) module implements the support vector machines algorithm. It is a supervised classification method that computes a maximal separating hyperplane between the expression vectors of different classes or phenotypes. Given microarray data with n markers per sample, SVM outputs a hyperplane,W, which can be thought of as a vector with n components each corresponding to the expression of a particular marker. Loosely speaking, assuming that the expression values of each marker have similar ranges, the absolute magnitude of each element in W determines its importance in classifying a sample.
Parameters
The SVM module has no settable parameters for the computation.
GenePattern Server Settings
To run GenePattern components, a GenePattern account is required.
- Protocol: HTTP or HTTPS, depending on the server being used.
- Host: URL of a GenePattern server.
- Port: Port at which the GenePattern server is located on the Host machine.
- Username: A valid user name on the specified GenePattern server.
- Password: A password, if required by the specified server.
References:
- R. Rifkin, S. Mukherjee, P. Tamayo, S. Ramaswamy, C-H Yeang, M. Angelo, M. Reich, T. Poggio, E.S. Lander, T.R. Golub, J.P. Mesirov, An Analytical Method for Multiclass Molecular Cancer Classification, SIAM Review, 45:4, (2003).
- T. Evgeniou, M. Pontil, T. Poggio, Regularization networks and support vector machines, Adv. Comput. Math., 13 (2000), pp. 1-50.
- V. Vapnik, Statistical Learning Theory, Wiley, New York, 1998.