Normalization
Overview
Normalization is used to transform data in preparation for analysis. Many normalizers are oriented towards decreasing the effects of systematic differences across a set of microarrays, aiding in cross-microarray comparisons. A log transformation of the data can be used to improve its statistical distribution. geWorkbench offers a selection of pluggable normalization components (see list below). The Normalizer panel is located in the Commands/ Analysis area in the lower right side of the application.</p>
In geWorkbench, normalization alters the loaded dataset and is not reversible; the original is not retained. Normalization operations do not respect any marker or array sets that may be activated; normalization always acts on the entire data set.
The Missing Value Computation normalizer can be used to replace missing values with imputed values. Note that some analysis routines also will optionally compute replacements for missing values if needed.
Data preparation methods such as RMA and GCRMA are not directly available in geWorkbench. Affymetrix CEL files can be processed externally to geWorkbench using a program such as RMAExpress or in R/Bioconductor and then the processed data imported into geWorkbench.
A short introduction to various methods of Affymetrix data preparation is available at Affymetrix Preprocessing.
Available Normalizers=
geWorkbench comes with the following normalization routines installed:
| Normalizer | Description | |
|---|---|---|
| Missing value computation | Replaces every missing value with either the mean value of that marker across all microarrays or with the mean measurement of all markers in the microarray where the missing value is observed | |
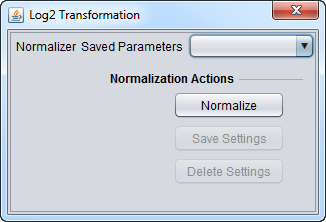
| Log2 Transformation | Applies a log2 transformation to all measurements in a microarray. | |
| Threshold Normalizer | All data points whose value is less than (or greater than) a user-specified minimum (maximum) value are raised (reduced) to that minimum (maximum) value | |
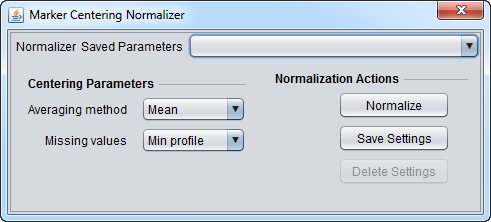
| Marker-based centering | Subtracts the mean or median measurement of a marker profile from every measurement in the profile | |
| Array-based centering | Subtracts the mean or median measurement of a microarray from every measurement in that microarray | |
| Mean-variance normalizer | For every marker profile, the mean measurement of the entire profile is subtracted from each measurement in the profile and the resulting value is divided by the standard deviation | |
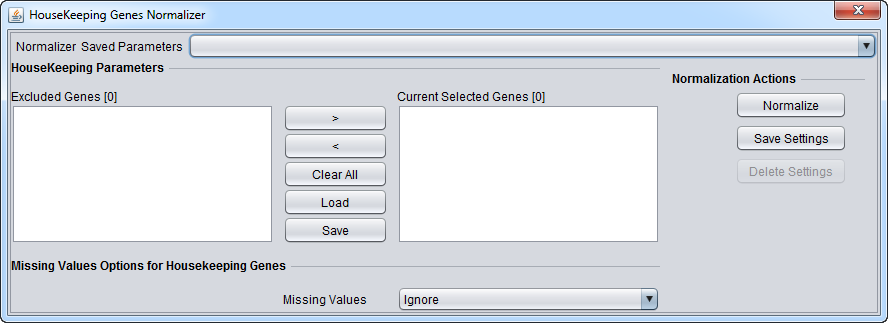
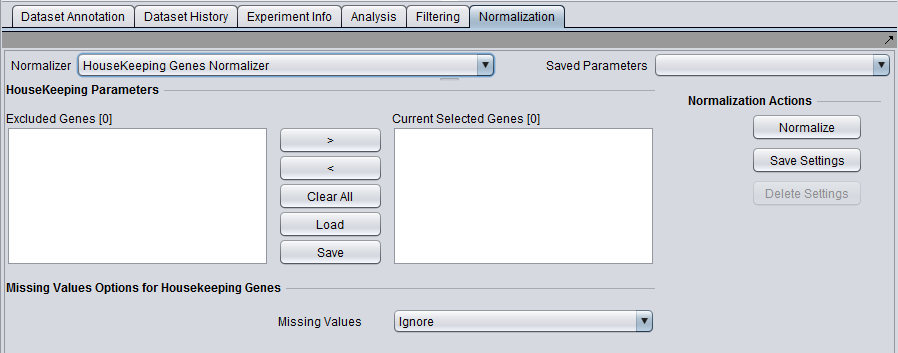
| House-keeping genes normalizer | Normalize all values such that the averaged expression value of specified house-keeping markers is the same on each microarray. | |
| Quantile Normalizer | Adjusts expression values so that the distribution of values is the same on each microarray, though which marker has which value varies. |