User:Daly
Contents
- 1 Overview
- 2 Tutorials
- 2.1 Getting Started
- 2.2 Loading Data
- 2.3 Working with Marker and Phenotype Panels
- 2.4 Visualize Gene Expression
- 2.5 Filter and Normalize Data
- 2.6 Clustering Gene Expression Data
- 2.7 Differential Expression
- 2.8 Regulatory Network
- 2.9 Integrated Annotation Information
- 2.10 Enrichment Analysis
- 2.11 Sequence Analysis
- 2.12 Pattern Discovery
- 2.13 Promoter Analysis
Overview
'(who uses this , why/ what for? background )
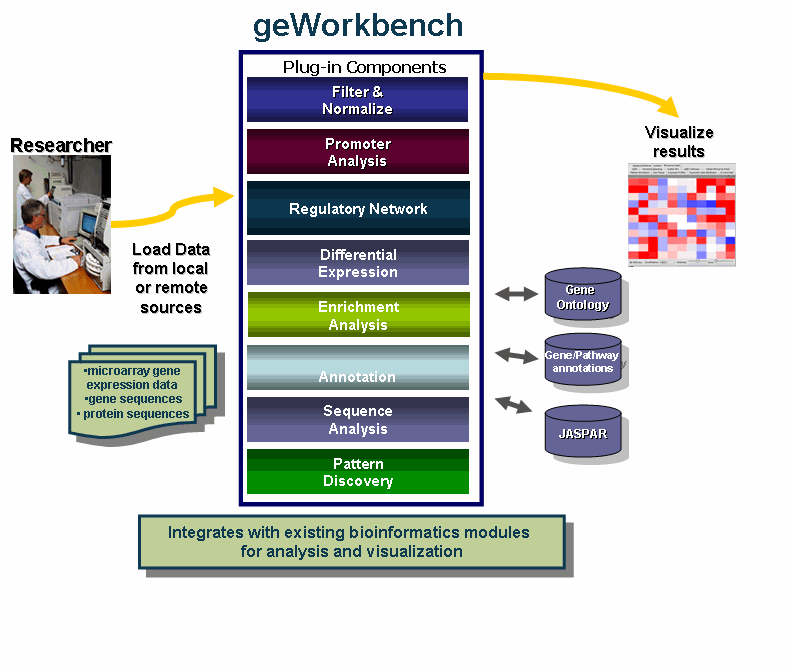
geWorkbench is an open-source bioinformatics platform that offers a comprehensive and extendible collection of tools for the management, analysis, visualization and annotation of biomedical data.
Benefits include:
- Integration with existing bioinformatics modules for analysis and visualization.
- Support for a variety of genomic data including microarrays, sequences, pathways, networks, alignments and phenotypes.
- Access to remote servers and clusters for the performance of computationally intensive calculations.
- Accesses analyses with biological annotations from the National Cancer Institute.
- Flexible import options: Allows user to merge files from various sources.
- Community: decribe this aspect
- Insert developer benefit ( plugin)
Tutorials
The following { insert description)
Getting Started
- Starting the application
- GUI elements
- Panels
- Navigation
Loading Data
- Data formats
Working with Marker and Phenotype Panels
Creating panels
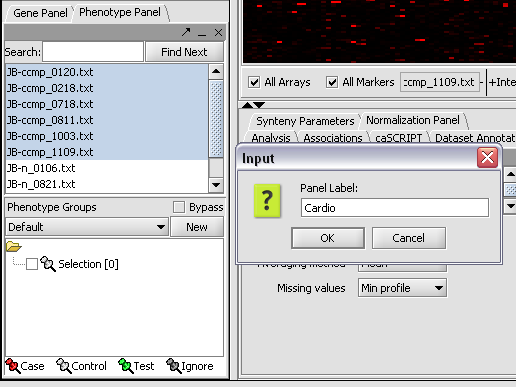
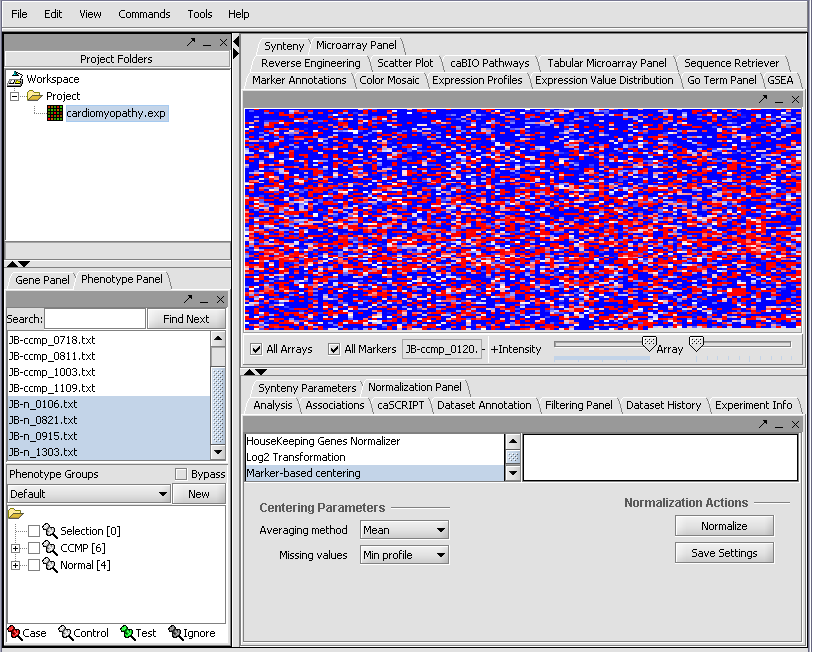
We can now assign phenotypes to each chip. We will place the phenotypes in the default group, however you can create new phenotype groups by pushing the New button on the Phenotype Panel at lower left.
Here we select and label arrays in the Phenotype Panel which contain samples from the congestive cardiomyopathy disease state...
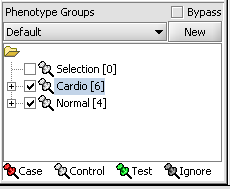
Next, we can similarly label the remaining arrays as "Normal". We have also checked boxes to indicate that these groups of arrays are "Active". Various analysis and visualization components can be set to only use/display activated arrays or markers.
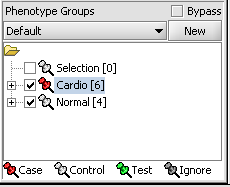
For statistical tests such as the t-test the Case and Control groups can be specified. This is done by left-clicking on the thumb-tack icon in front of the phenotype name. Here we are specifying the disease arrays as the "Case". The remaining "Normal" arrays are by default labeled control.
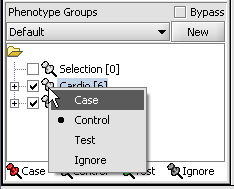
A red thumbtack indicates the arrays have been specified as "Case".
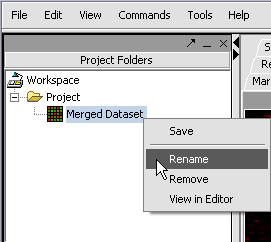
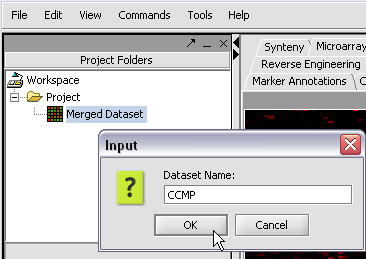
We can also rename the merged dataset by clicking on its entry in the Project Panel.
Here we will call it CCMP.
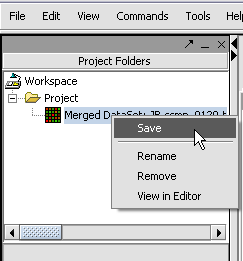
With the datasets merged, classified and named, we can save the dataset for future use. We will call it "cardiomyopathy.exp" (.exp is the default extension for the geWorkbench matrix file type).
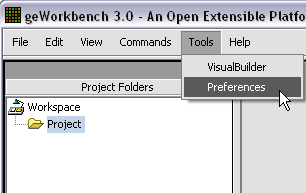
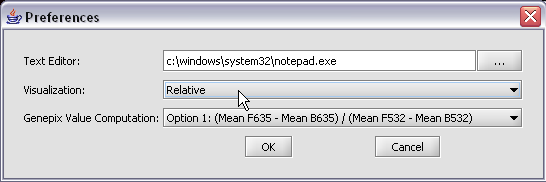
The default display of microarray data is an absolute display. We can change it to a relative display by selecting Tools:Preferences from the top menubar. We have removed the dataset so that we can read it back in using the new preferences.
Here we select the relative display type.
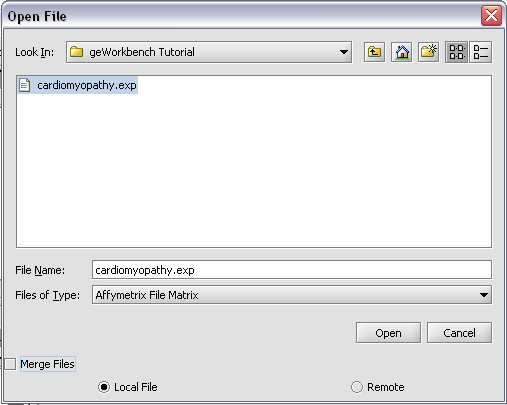
Returning to the Open File dialog as we before by right-clicking on the project entry, we will select the "cardiomyopathy.exp" file we previously saved...
Resulting in the following colorful display of the array data for the first array....
Visualize Gene Expression
Filter and Normalize Data
Clustering Gene Expression Data
- Hierarchical Clustering
- Self Organizing Map (SOM)
Differential Expression
- T Test
- Multi Test
- Volcano Plot
- Color Mosaic
Regulatory Network
- Reverse Engineering
- Cytoscape
Integrated Annotation Information
Enrichment Analysis
- Go Term
- Go Miner
Sequence Analysis
- Sequence Retrieval
- Sequence Homology Analysis
- Blast
- Other
Pattern Discovery
- Position Histogram