Difference between revisions of "CeRNA Query"
| Line 2: | Line 2: | ||
The interactions were computed based on TCGA expression data sets. | The interactions were computed based on TCGA expression data sets. | ||
| + | |||
| + | Datasets are available for Glioblastoma (GBM), Ovarian (OV), Breast (BRCA), and Prostate (PRAD) cancer datasets. | ||
Revision as of 15:33, 2 August 2013
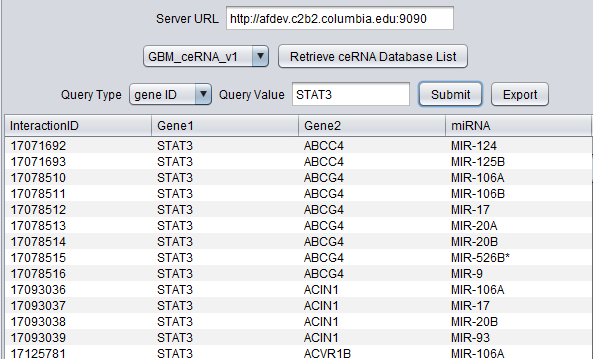
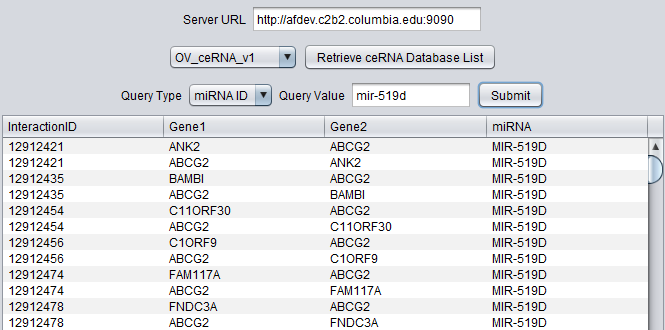
This component provides query access to a precomputed database of competitive endogenous RNA (ceRNA) interactions, also called "sponge" interactions. These interactions were predicted using the Hermes algorithm (Sumazin et al., 2011) with subsequent improvements (submitted). Hermes is used to predict the intersection set of miRNAs which bind to both of two different RNAs. Variation in the level of one of the RNA species may affect the level of the second RNA via its effects on the availability of the various miRNAs, providing a regulatory mechanism for gene expression.
The interactions were computed based on TCGA expression data sets.
Datasets are available for Glioblastoma (GBM), Ovarian (OV), Breast (BRCA), and Prostate (PRAD) cancer datasets.
Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J, Califano A. (2011) An extensive microRNA-mediated network of RNA-RNA interactions regulates established oncogenic pathways in glioblastoma. Cell 147(2):370-81. doi: 10.1016/j.cell.2011.09.041.