Difference between revisions of "Color Mosaic"
| Line 1: | Line 1: | ||
| − | == | + | ==Overview== |
The Color Mosaic component displays expression values for all active arrays and markers using a "heat map" colored by either an absolute or relative color scheme. | The Color Mosaic component displays expression values for all active arrays and markers using a "heat map" colored by either an absolute or relative color scheme. | ||
| − | + | ==Controls== | |
| − | + | ===Display==== | |
Pushing the '''Display''' button causes the view to be drawn or redrawn. | Pushing the '''Display''' button causes the view to be drawn or redrawn. | ||
| − | + | ===Accession=== | |
If the '''Accession''' button is active, probeset names will be displayed at right for each marker. | If the '''Accession''' button is active, probeset names will be displayed at right for each marker. | ||
| − | + | ===Label=== | |
If the '''Label''' button is active, gene names will be displayed at right for each marker. | If the '''Label''' button is active, gene names will be displayed at right for each marker. | ||
| − | + | ===Export data=== | |
| − | + | ===Sort=== | |
| − | + | ===Bulb icon=== | |
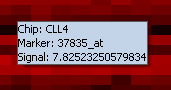
Activates a mouse-over tool tip. When the cursor is placed over a point on the color mosaic display, the array, marker, and expression value are displayed. | Activates a mouse-over tool tip. When the cursor is placed over a point on the color mosaic display, the array, marker, and expression value are displayed. | ||
| Line 28: | Line 28: | ||
| − | + | ===Intensity slider=== | |
The slider increases or decreases the thresholds used to define the color codings, as explained below in the Color Key section. | The slider increases or decreases the thresholds used to define the color codings, as explained below in the Color Key section. | ||
| − | + | ===Color key=== | |
The Color key shows the range of colors available in the currently set Visualization preference (Menu->Tools->Preferences->Visualization). There are two available color schemes: '''Absolute''' and '''Relative'''. In each, the lowest and highest expression values correspond to the most saturated colors, and values closer to the middle are indicated by shading into a third color. | The Color key shows the range of colors available in the currently set Visualization preference (Menu->Tools->Preferences->Visualization). There are two available color schemes: '''Absolute''' and '''Relative'''. In each, the lowest and highest expression values correspond to the most saturated colors, and values closer to the middle are indicated by shading into a third color. | ||
| − | + | ====Absolute==== | |
Colors are calculated on an absolute scale centered about zero (when the Intensity Slider is in the middle, neutral position). Values shade between the following: | Colors are calculated on an absolute scale centered about zero (when the Intensity Slider is in the middle, neutral position). Values shade between the following: | ||
| Line 41: | Line 41: | ||
* Green - negative values. | * Green - negative values. | ||
| − | + | ====Relative==== | |
Colors are calculated relative to the mean '''for each marker''' (when the Intensity Slider is in the middle, neutral position). Expression value colors are directly comparable only within a particular marker. | Colors are calculated relative to the mean '''for each marker''' (when the Intensity Slider is in the middle, neutral position). Expression value colors are directly comparable only within a particular marker. | ||
| Line 48: | Line 48: | ||
* Blue - values less than the mean. | * Blue - values less than the mean. | ||
| − | + | ===Gene Height and Gene Width=== | |
Change the size (number of pixels) assigned to each entry (tile) in the color mosaic. The also changes the size of the associated labels. | Change the size (number of pixels) assigned to each entry (tile) in the color mosaic. The also changes the size of the associated labels. | ||
| − | + | ==Actions== | |
| − | + | ===Clicking on an entry=== | |
Clicking on any location (tile) in the mosaic will highlight the corresponding gene probe in the Markers component, and the corresponding array in the Arrays/Phenotypes component. | Clicking on any location (tile) in the mosaic will highlight the corresponding gene probe in the Markers component, and the corresponding array in the Arrays/Phenotypes component. | ||
Revision as of 17:19, 28 July 2009
Contents
Overview
The Color Mosaic component displays expression values for all active arrays and markers using a "heat map" colored by either an absolute or relative color scheme.
Controls
Display=
Pushing the Display button causes the view to be drawn or redrawn.
Accession
If the Accession button is active, probeset names will be displayed at right for each marker.
Label
If the Label button is active, gene names will be displayed at right for each marker.
Export data
Sort
Bulb icon
Activates a mouse-over tool tip. When the cursor is placed over a point on the color mosaic display, the array, marker, and expression value are displayed.
Intensity slider
The slider increases or decreases the thresholds used to define the color codings, as explained below in the Color Key section.
Color key
The Color key shows the range of colors available in the currently set Visualization preference (Menu->Tools->Preferences->Visualization). There are two available color schemes: Absolute and Relative. In each, the lowest and highest expression values correspond to the most saturated colors, and values closer to the middle are indicated by shading into a third color.
Absolute
Colors are calculated on an absolute scale centered about zero (when the Intensity Slider is in the middle, neutral position). Values shade between the following:
- Black - zero.
- Red - positive values.
- Green - negative values.
Relative
Colors are calculated relative to the mean for each marker (when the Intensity Slider is in the middle, neutral position). Expression value colors are directly comparable only within a particular marker.
- White - mean value for marker.
- Red - values greater than the mean.
- Blue - values less than the mean.
Gene Height and Gene Width
Change the size (number of pixels) assigned to each entry (tile) in the color mosaic. The also changes the size of the associated labels.
Actions
Clicking on an entry
Clicking on any location (tile) in the mosaic will highlight the corresponding gene probe in the Markers component, and the corresponding array in the Arrays/Phenotypes component.