Difference between revisions of "Cytoscape Network Viewer"
| Line 1: | Line 1: | ||
| + | Cytoscape (www.cytoscape.org) is a sophisticated network and pathway visualization tool that has been incorporated into geWorkbench as a component. Within geWorkbench, Cytoscape is used to depict putative interaction networks, for example as created from running ARACNe or a Cellular Network Knowledgebase query. Both of these tools return "adjacency matrices", that is, interaction networks, to the Project folders component. Currently, Cytoscape version 2.4.7 used in geWorkbench. | ||
| + | |||
| + | Cytoscape has been integrated into geWorkbench in such a way that it can communicate in both directions with the Markers component. | ||
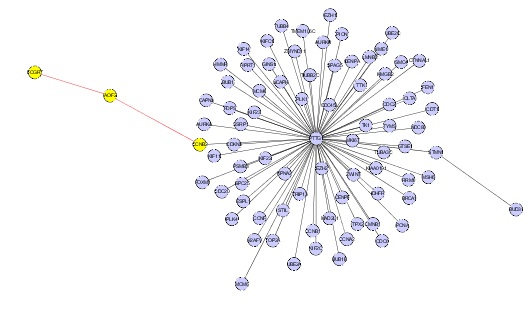
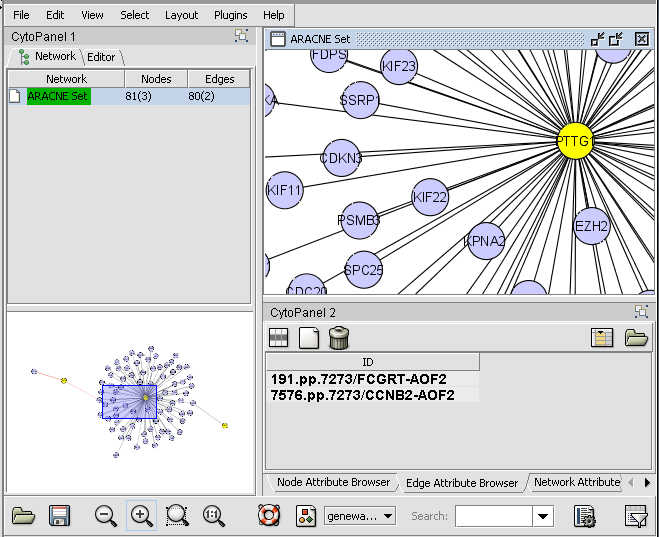
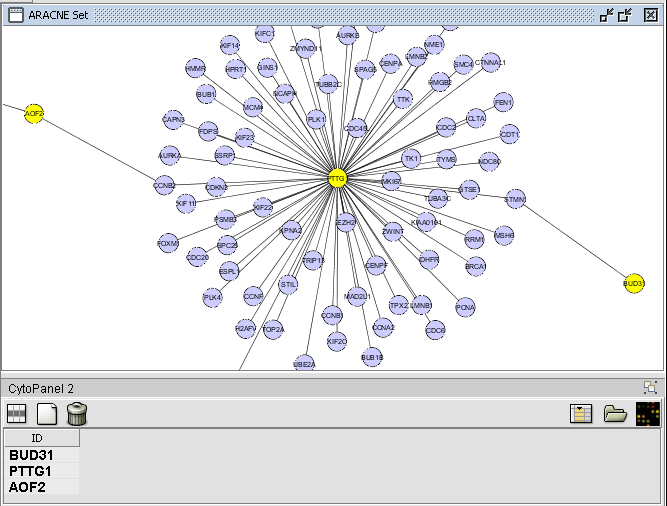
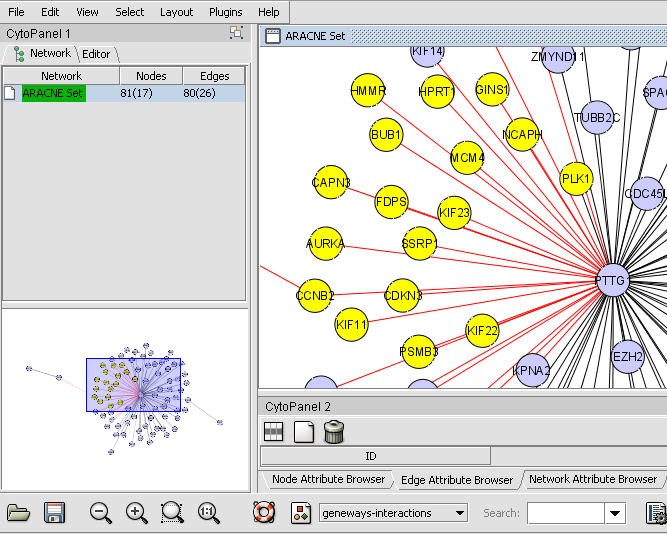
| + | # Nodes in a Cytoscape network can be selected indivudually or by drawing a selection box around them. This will result in the selected nodes being placed into the "Cytoscape selection" set in the Markers component. | ||
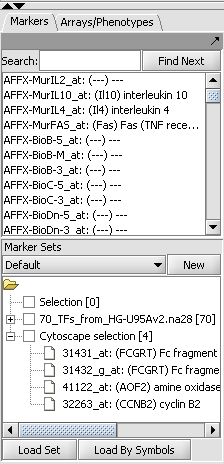
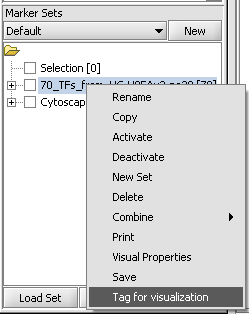
| + | # A set of markers in the Markers component can be labeled with the "tag for visualization" property, which will project that set onto the network depicted in Cytoscape. Those markers in the intersection of the tagged set and the network display will be hightlighted in yellow. | ||
| + | |||
| + | These actions are illustrated in the following examples. The network diagram depicted is that calculated using ARACNe in the [http://wiki.c2b2.columbia.edu/workbench/index.php/Tutorial_-_ARACNE ARACNe tutorial]. | ||
| − | |||
Revision as of 11:01, 14 July 2009
Cytoscape (www.cytoscape.org) is a sophisticated network and pathway visualization tool that has been incorporated into geWorkbench as a component. Within geWorkbench, Cytoscape is used to depict putative interaction networks, for example as created from running ARACNe or a Cellular Network Knowledgebase query. Both of these tools return "adjacency matrices", that is, interaction networks, to the Project folders component. Currently, Cytoscape version 2.4.7 used in geWorkbench.
Cytoscape has been integrated into geWorkbench in such a way that it can communicate in both directions with the Markers component.
- Nodes in a Cytoscape network can be selected indivudually or by drawing a selection box around them. This will result in the selected nodes being placed into the "Cytoscape selection" set in the Markers component.
- A set of markers in the Markers component can be labeled with the "tag for visualization" property, which will project that set onto the network depicted in Cytoscape. Those markers in the intersection of the tagged set and the network display will be hightlighted in yellow.
These actions are illustrated in the following examples. The network diagram depicted is that calculated using ARACNe in the ARACNe tutorial.