Cytoscape Network Viewer
Contents
- 1 Overview
- 2 Text Viewer for Large Networks
- 3 Gene Level Summarization of Networks
- 4 Layout of the Cytoscape component
- 5 Changes in Cytoscape 2.8
- 6 Selecting Nodes and Edges
- 7 Options for selected nodes
- 8 Making bulk changes to node or edge properties
- 9 Projecting marker sets onto Cytoscape
- 10 Altering the view in Cytoscape
- 11 Network commands
- 12 Edges colored by interaction type
- 13 Cytoscape VizMapper tab - set shapes and color
- 14 Saving Changes to Network Attributes
- 15 geWorkbench Network Operations
- 16 References
Overview
Cytoscape (www.cytoscape.org) is a sophisticated network and pathway visualization tool that has been incorporated into geWorkbench as a component. Within geWorkbench, Cytoscape is used to depict putative interaction networks, for example as created from running ARACNe or a Cellular Network Knowledge Base query. Both of these tools return "adjacency matrices", that is, interaction networks, to the Workspace.
Cytoscape has been integrated into geWorkbench in such a way that it can communicate in both directions with the Markers component.
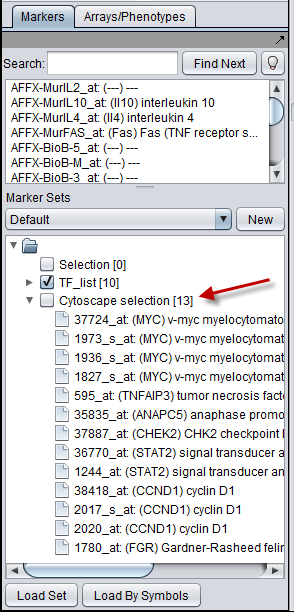
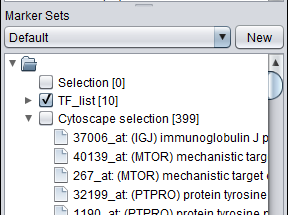
- Nodes in a Cytoscape network can be selected individually or by drawing a selection box around them. This will result in the selected nodes being placed into the "Cytoscape selection" set in the Markers component.
- A set of markers in the Markers component can be labeled with the "tag for visualization" property, which will project that set onto the network depicted in Cytoscape. Those markers in the intersection of the tagged set and the network display will be highlighted in yellow.
The use of Cytoscape, and its interactions with geWorkbench, are described in the following sections. The figures shown in this chapter were generated using the query results obtained in the Cellular_Networks_KnowledgeBase chapter.
First we will describe the layout of the Cytoscape graphical interface.
Text Viewer for Large Networks
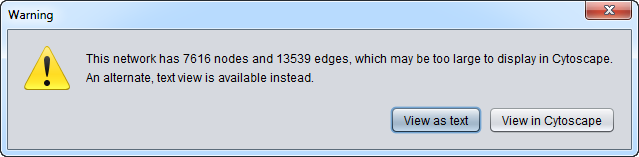
There is a limit to the size of networks that Cytoscape can be used to visualize in geWorkbench. This limit will depend on the amount of memory allocated to geWorkbench. Viewing too large a network can cause geWorkbench to run out of memory and stop responding.
As of release 2.2.1, if the user attempts to view a large network in Cytocape, he or she will be offered the option to instead view the network in a text viewer.
The default number for triggering this choice is that the sum of nodes (genes) and edges (interactions) is set to a very conservative value of 5000. This number can be altered by changing a setting in the Tools->Preferences menu. The setting is called "Soft Limit on Cytoscape Networks Objects (nodes+edges)".
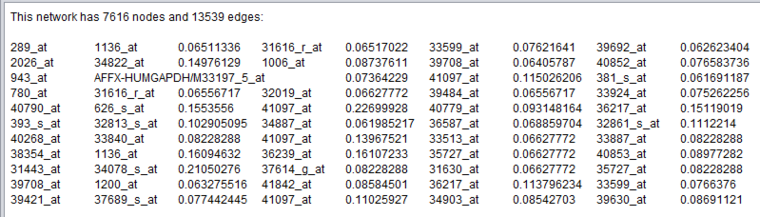
Below is an example of a portion of an ARACNe adjacency matrix node displayed using the text viewer.
Gene Level Summarization of Networks
If the network to be viewed is represented by e.g. Affymetrix probesets, and if an appropriate annotation file has been loaded, then geWorkbench will summarize the network at the gene level before displaying it using Cytoscape. A gene may be represented by more than one probeset. All edges connected to probesets representing one gene are assigned to a single gene node. This may cause the number of nodes and edges displayed in Cytoscape to be less than those in the original probeset-level network. You can see the number of nodes and edges in the original network by hovering the mouse cursor over the network data node in the Workspace.
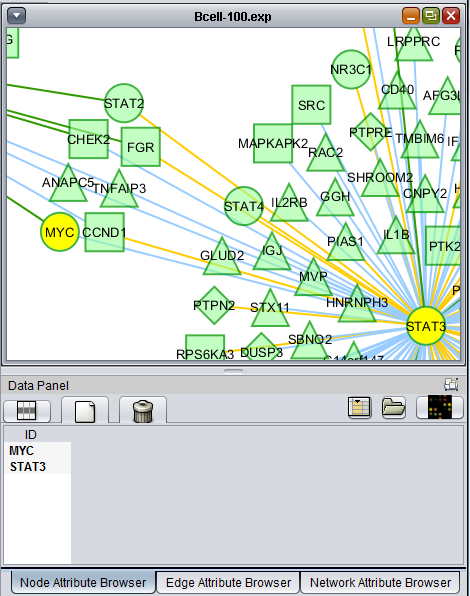
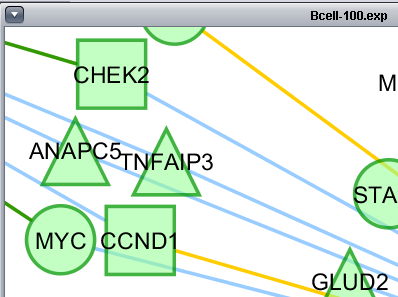
Layout of the Cytoscape component
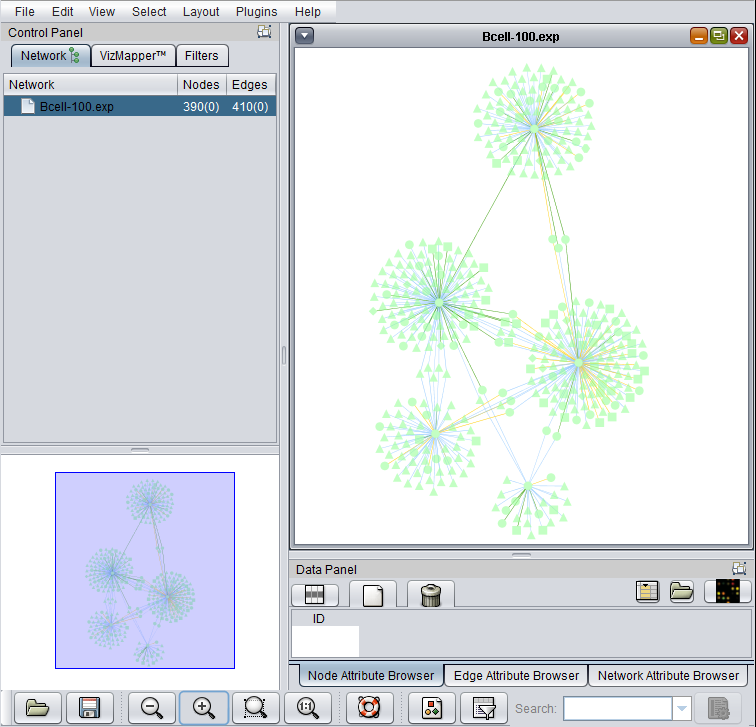
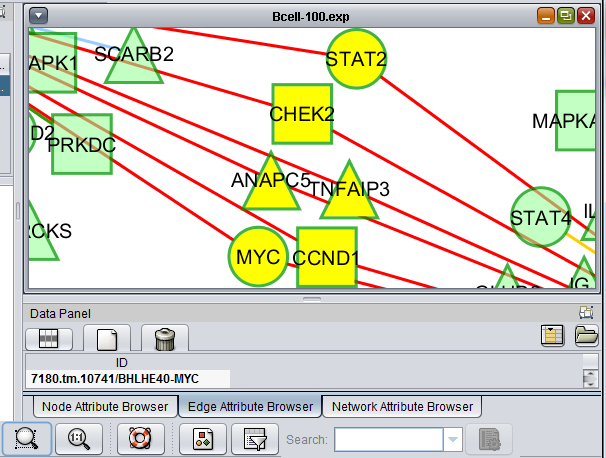
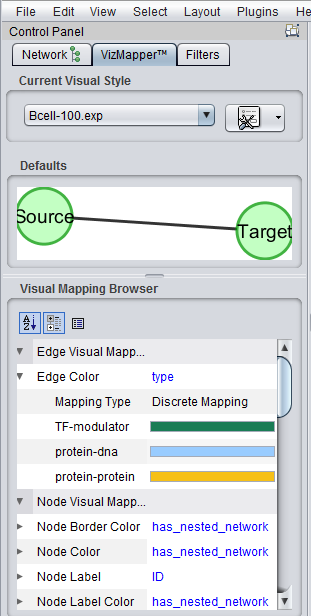
The Cytoscape component has 4 main areas:
- At upper left is a list of networks that have been loaded into Cytoscape, showing the network name and the number of nodes and edges. In addition, the numbers in parentheses show the numbers of nodes and edges which have been highlighted (selected) in the network depiction.
- At upper right is the main network depiction pane. Gene or protein symbols/names will be depicted if available. However, if for example an Affymetrix microarray dataset was read in but no annotation file was associated with it, then only probeset names will appear. Selected nodes are depicted in yellow and are returned to the "Cytoscape selection" set in the Markers component. Selected edges are depicted in red.
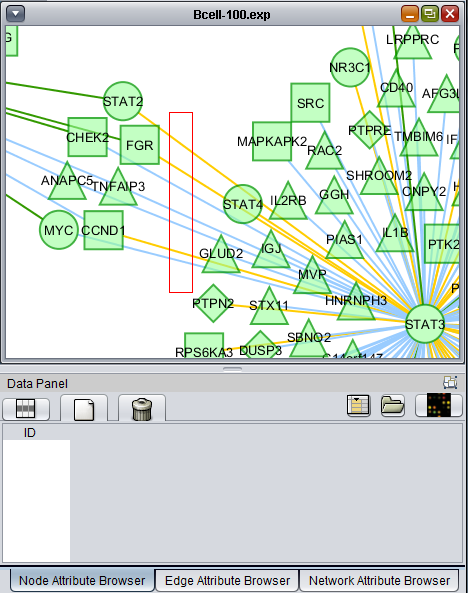
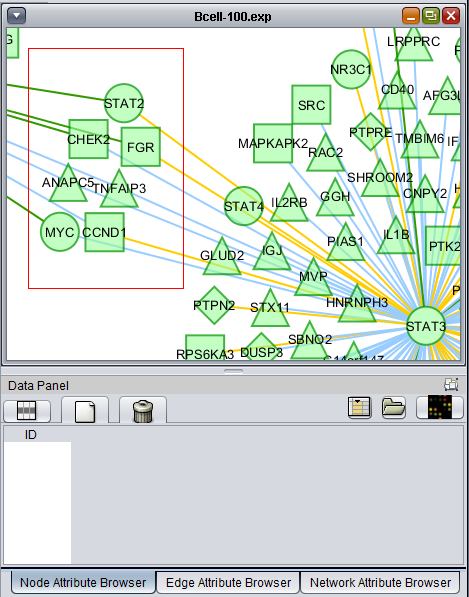
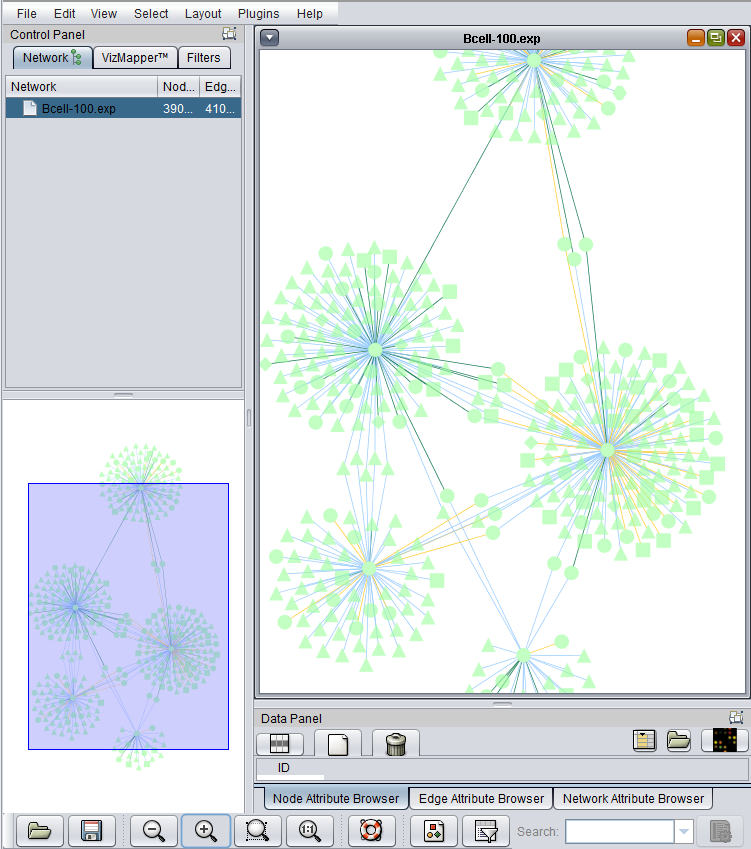
- At lower left is a navigation tool which shows the entire network and the location and size (purple rectangle) of the current viewing pane. The purple viewing pane can be moved about the network as desired. This is done by left-clicking with the mouse in the purple rectangle and moving it.
- At lower right, nodes or edges that have been selected in the network display will appear in the Data Panel.
Note - the small controls at upper right of the "Control Panel" and the "Data Panel", which are used to "float" these panels free of the application, have been disabled in geWorkbench.
Changes in Cytoscape 2.8
With geWorkbench v2.3.0, the version of Cytoscape was updated to 2.8. There are several differences in the Cytoscape graphical interface, compared with that depicted in the remainder this chapter.
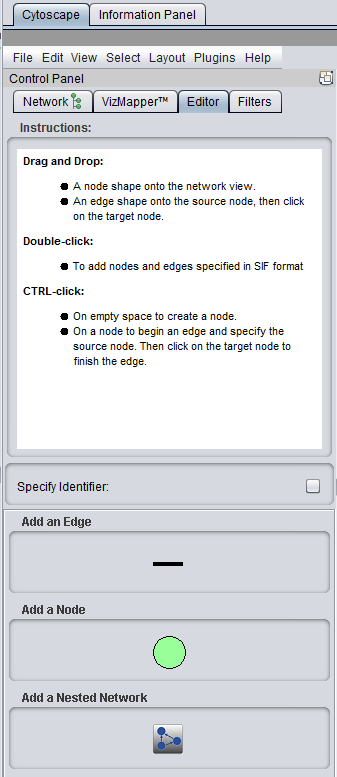
Control Panel - Editor Pane added.
Toolbar - new tool icons added.
Data Panel - new tool icons added.
The icons on the left side of the data panel represent, from left to right:
- Select Attribute
- Create New Attribute
- Select All Attributes
- Unselect All Attributes
- Delete Attributes
The icons on the right side of the data panel represent, from left to right:
- Attribute Batch Editor
- Function Builder
- Import Attributes from File
- Import Expression Matrix Data
Selecting Nodes and Edges
This section depicts the network generated in the Cellular_Networks_KnowledgeBase chapter.
Selecting interactions (edges)
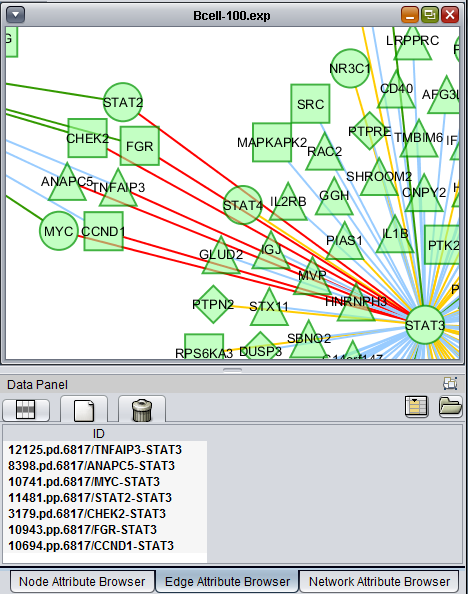
Using the mouse, a group of edges can be selected. Hold down the mouse button while drawing a box on the screen around the desired edges. The box will appear in red.
The list of selected edges is displayed in a list below the graph, in a tab titled "Edge Attribute Browser".
Selecting Markers/Genes (nodes)
In geWorkbench, the nodes may use as their primary identifier either the gene name or marker id, depending on the origin of the network being displayed. Aadditional display attributes can be added in the Node Attribute Browser.
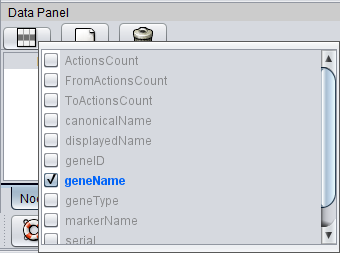
Here we click on the leftmost icon, "Select Attributes", and add the Gene Name.
- Individual nodes and/or edges can be selected in Cytoscape by clicking on them with the mouse.
- To select multiple nodes or edges, hold down the Shift key while making the selection.
- Alternatively, a selection box can be drawn around both nodes and edges by left-clicking in the network diagram and selecting the desired targets.
The Data Panel
Left- or right-clicking on markers displayed in the Data Panel Node Attribute Browser will produce link-out choices to annotation pages. For a few annotation sources, such as UniProt, only one or the other (right or left click) may successfully link out to the web page. (Mantis issues #2715, #2716).
The "Cytoscape_Selection" marker set
Selecting nodes in Cytoscape will cause matching markers to appear in the "Cytoscape_Selection" set in the Markers component. The exact way in which markers are transfered depends on the origin of the network. The selection is dynamic - changing the selected nodes will change the contents of the "Cytoscape_Selection" set.
To make a permanent copy of a particular set of dynamically chosen markers in the "Cytoscape Selection" subset, right-click on it and select "Copy".
- Network represented by gene symbols - selecting a node will cause all markers annotated to that gene to appear in the "Cytoscape_Selection" set in the Markers component. This is true even for genes that are not the first associated with a given probeset in the annotation file.
- Network represented by marker ids - selecting a node will cause only the selected marker to appear in the Markers component.
Selecting both nodes and edges
A selection box, described above for selecting edges, can also be drawn to include both nodes and edges at the same time.
The selected nodes and edges will be listed in the Node Attribute Browser and Edge Attribute Browser lists below the network diagram.
Options for selected nodes
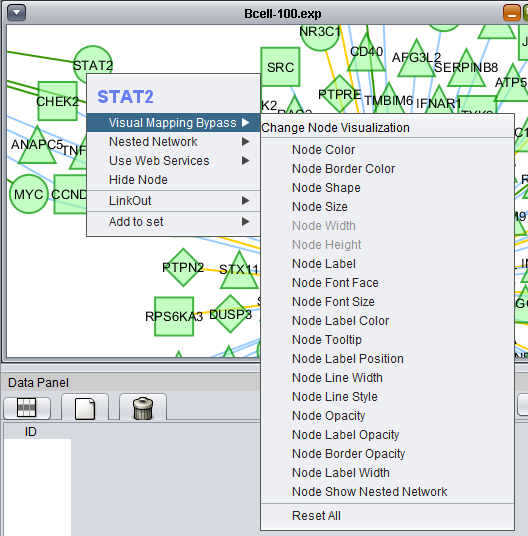
Right-clicking on a particular node in the network graph brings up a menu with a number of options describe below.
Visual Mapping Bypass
From the Visual Mapping Bypass menu, any attribute of the node such as color and shape can be altered. This appears to only work on one node at a time.
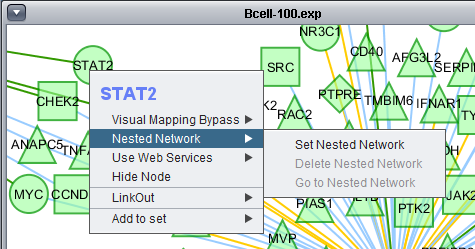
Nested Network
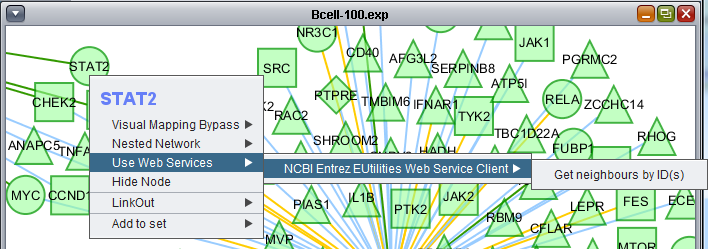
Use Web Services
Please see the Ctyoscape manual.
Hide Node
Selecting "Hide Node" will cause any selected nodes and all their edges to disappear.
Hidden nodes can be made visible again by right-clicking on the background and selecting "Restore network".
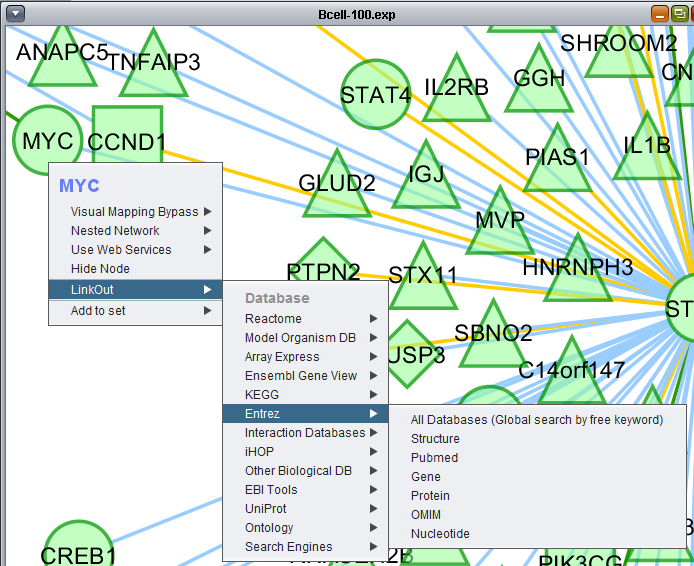
LinkOut
This menu option provides hyperlinks to a number of external sources of gene annotation.
Note - UniProt Workaround - The UniProt ID link-outs may not work directly. However, you can highlight a node of interest, and then, if present, select its UniProt ID node in the Cytoscape Data Panel. Depending on the specific situation, either right- or left-clicking on a link there may produce a working link-out. (Mantis issues #2715, #2716).
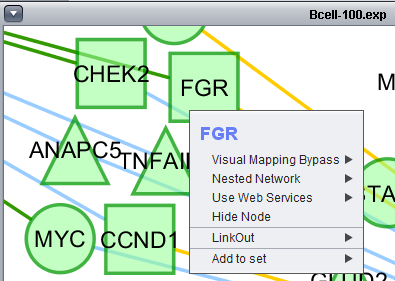
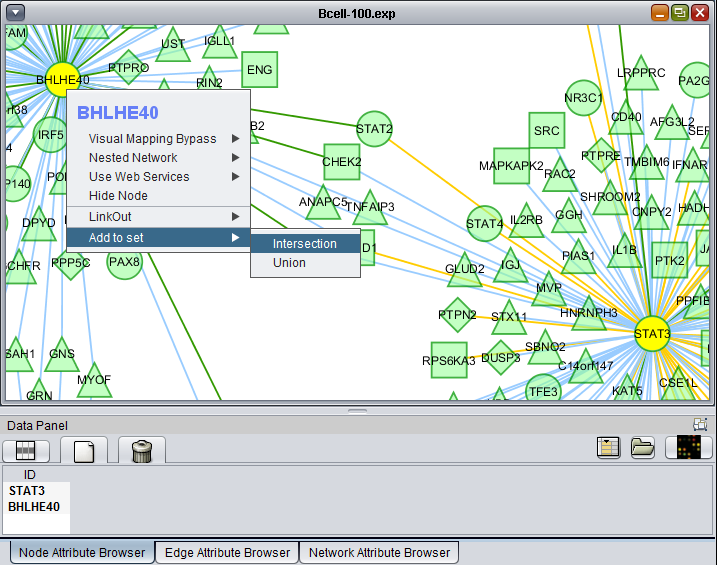
Add to set
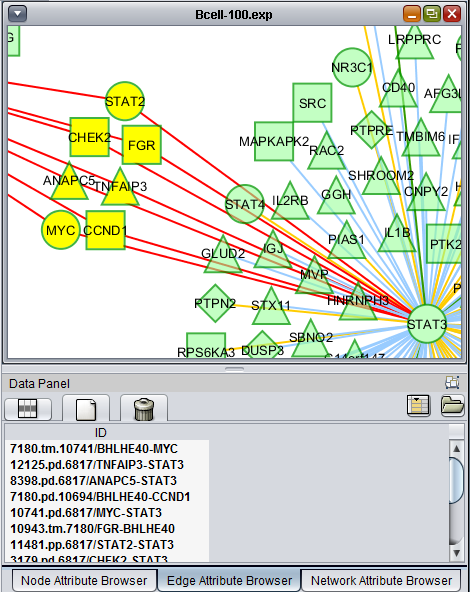
If one or more graph nodes have been selected (highlighted in yellow in figure below), the markers they directly interact with (via edges) can be copied to the default "Cytoscape selection" subset in the Markers component at lower left in the geWorkbench graphical interface.
Two options are available under "Add to Set". These are
- Intersection - find the set of markers that have interactions (edges) with ALL selected nodes. Such markers are placed into the Markers component, in the "Cytoscape selection" subset.
- Union - find the set of markers that have interactions (edges) with ANY of the selected nodes. Such markers are placed into the Markers component, in the "Cytoscape selection" subset.
This image shows the intersection set of markers for the two selected genes. There are 30 markers in the intersection:
This image shows the union set of markers for the two selected genes. There are 251 markers in the union.
Note - For each gene included in the UNION or INTERSECTION, there may be more than one marker associated with it. If so, all markers belonging to a particular gene in a UNION or INTERSECTION result will be returned to to the Markers component "Cytoscape Selection" set.
Making bulk changes to node or edge properties
Using the Visual Mapping Bypass menu appears to affect only one node or edge at a time. However, there is a way to change the properties of multiple nodes or edges at the same time. The user may wish to consult the Cytoscape manual, but here is one method that works:
- These steps assume you have already generated your network and it is displayed in Cytoscape.
- In the Data Panel, select the Node or Edge Attribute Browser, depending on the type of object for which you wish to change a property.
- For this example, we set node colors for multiple nodes.
- Find the icon for "Select Attributes" and push it. The available attributes are shown. If there is no attribute defined for what you wish to change,
- create a new one by finding and pushing the "Create New Attribute" button.
- Create the attribute as type "String" and give it a meaningful name, such as MyNodeColor.
- Select the set of nodes that you wish to color.
- Find the "Attribute Batch Editor" icon at right on the Data Panel and push it.
- In the dialog, choose "Set", then the name of your attribute, e.g. MyNodeColor, and then give the new value a name, such as "red_orange".
- You can add as many new values for the attribute as you like to different sets of nodes, e.g. "reddish", "blue_yellow", as desired.
- Go to the VizMapper tab and find the property you wish to map, e.g. Node Color.
- Click on the field just to the right and set it to the attribute you created, e.g. MyNodeColor.
- Choose Discrete Mapping and click on the setter field to the right of each value in turn.
- A color chooser will allow you to assign a color to each value label.
The nodes will now be colored using the values you have assigned.
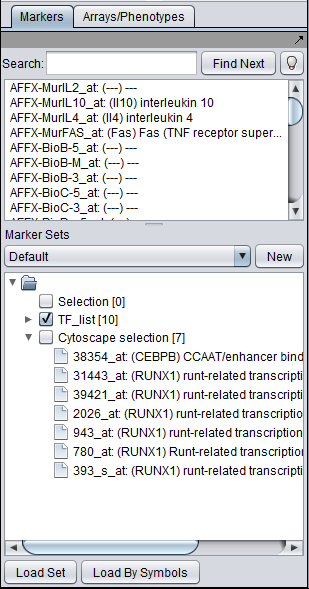
Projecting marker sets onto Cytoscape
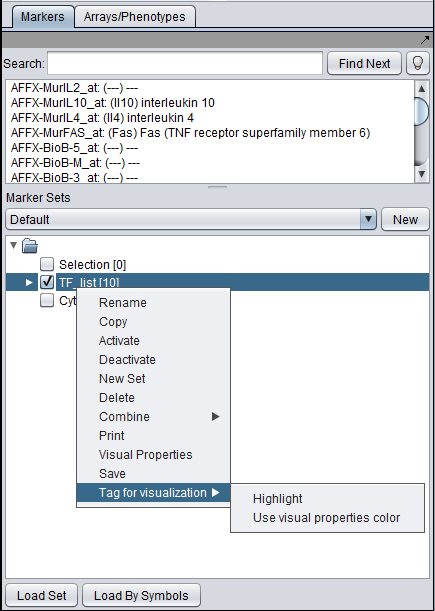
The diagram below illustrates projecting a set defined in the Markers component back onto the Cytoscape network diagram. In this case, the set of transcription factors originally used in to form the CNKB query which generated this example is labeled with "tag for visualization" by right-clicking on it and selecting this menu option.
Each gene or marker that is present at least once in the tagged set, and which is also present in the drawn network, is highlighted in yellow in the network display.
Altering the view in Cytoscape
The use of the sliding viewpane at lower left to navigate about the main drawn network has already been mentioned - it can be grabbed and moved by left-clicking on it with the mouse.
There are several more controls arrayed about the lower edge of the Cytoscape component.
For control the network view:
![]() Open - zoom to display selected region.
Open - zoom to display selected region.
![]() 1:1 - zoom out to display all of current network.
1:1 - zoom out to display all of current network.
To illustrate the "display selected" function, below we select several nodes...
...and click ![]() to zoom in on this portion of the graph.
to zoom in on this portion of the graph.
Network commands
Right-clicking on a listed network in Cytoscape will bring up a menu with the following choices:
Edit Network Title
Edit the title of the network.
Create View
Recreate the network graphics.
Destroy View
Remove the network graphics.
Destroy Network
Completely remove the network from Cytoscape. Note that this does not remove the network adjacency matrix from the geWorkbench Workspace. The network can be recreated in Cytoscape by clicking on the appropriate adjacency matrix in the Workspace.
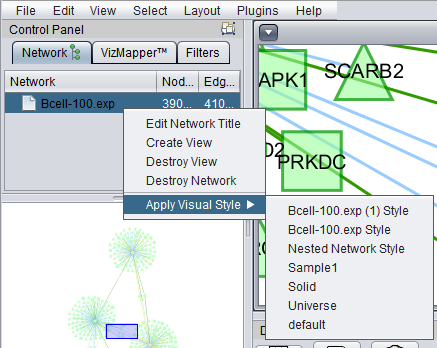
Apply Visual Style
The Apply Visual Style feature adds the option to apply user-defined styles to a network diagram, in addition to the various preset styles defined in Cytoscape. Edge colors are now included in these styles. Any changes that the user makes to the visual appearance of the network are saved in a new "style" entry with the name of the current dataset. These saved styles can then be applied to other datasets.
The default style is the "Nested Network Style".
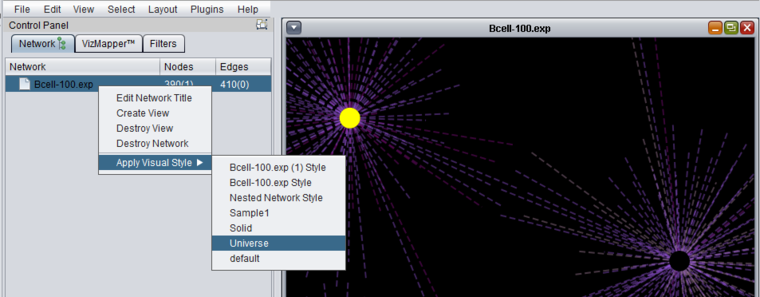
The figure below shows the result of applying the "Universe" style to a network where two nodes are selected (yellow).
Edges colored by interaction type
When the molecular interaction type is available, edges are colored according to the interaction type, e.g. Protein-DNA. Molecular interation types are stored in the Cellular Networks Knowledge Base (CNKB) and in SIF format network files, and are included when a network is created from either of those sources. The figure below shows the result of such a query against the CNKB.
The key for the assignment of colors to interaction types is described in the next section.
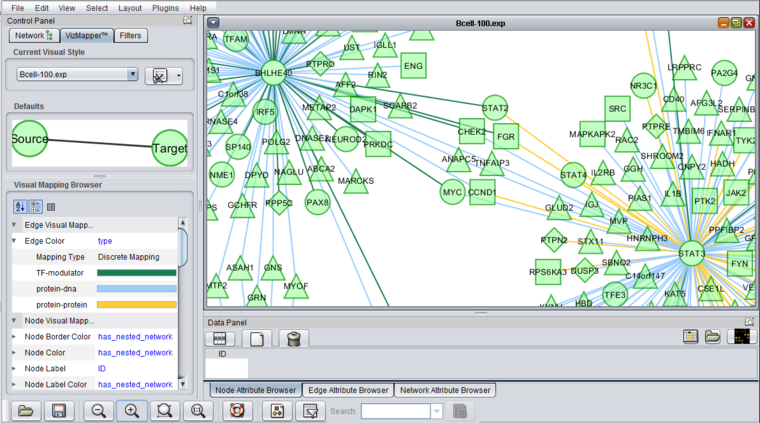
Cytoscape VizMapper tab - set shapes and color
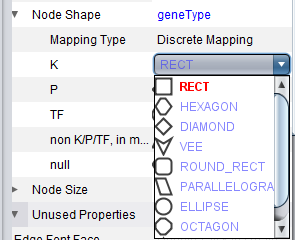
geWorkbench assigns shapes to genes in Cytoscape based on their activity, e.g. transcription factor, kinase, phosphatase etc.
The figure below shows a zoomed-in view of the network generated in the CNKB chapter. The VizMapper tab has been selected, and the properties list has been opened to the node shape key.
Clicking on a particular shape in the legend brings up a shape editor, with which the shape assignements can be changed.
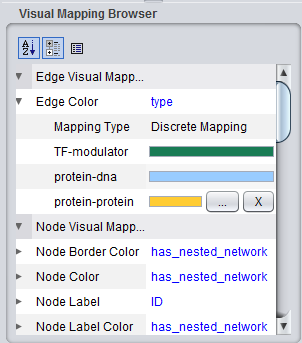
Similarly, when molecular interaction types are available, colors are assigned to the lines (termed edges) connecting genes, which represent their interactions. These edges are colored based on the type of interaction, e.g. Protein-DNA etc.
As currently implemented, colors are assigned randomly to each interaction type when geWorkbench starts up. There will thus be a different color palate with each invocation of geWorkbench. Within a single invocation of geWorkbench, the same color palate will be used for each network created.
The figure below shows the VizMapper edge color key.
The colors assigned to each interaction type can be edited by clicking on a particular color bar in the legend, as shown in the next figure. Two buttons appear, one for editing, labeled with "...", and one to delete the interaction color assignemt, "X". If color edit is chosen, a color chooser will appear.
Saving Changes to Network Attributes
You may be able to save to file and restore specific changes you have made to the network appearance using the Cytoscape Export and Import functions. This is not part of geWorkbench and is not covered further here.
geWorkbench Network Operations
geWorkbench adds four functions to those available in Cytoscape.
- Show t-test results
- Compute edge correlations.
- Create subnetwork.
- Restore network.
Show t-test results
Description
When a t-test has been run in geWorkbench, its results are placed in a child data node under the parent dataset in the Workspace. The results (in terms of a p-value calculated from the t-statistic for each marker) can be viewed as a "heat map" using the Color Mosaic component, or as a Volcano plot.
geWorkbench can also overlay t-test results onto a network displayed in Cytoscape. Genes which have a significant (above threshold) result in the t-test and which are present as nodes in the network can be colored according to the sign and magnitude of the marker t-statistic. Positive t-statistic values are shaded red, and negative values are shaded blue. The greater the absolute value of the t-statistic, the greater the intensity (deeper shade) of the color.
In cases where more than one marker (probeset) represents a gene, the result from the marker with the highest absolute t-value is used.
Note that in the Color Mosaic, the heat map displays the actual expression results for each marker and each array. Here, Cytoscape displays one summary value, the t-statistic, for each gene.
Prerequisites
- Only t-test result nodes belonging to the same parent microarray expression data node as the adjacency matrix being displayed in Cytoscape can be selected for overlay.
- The matching of markers to Cytoscape nodes is done using the Gene Symbol. For this reason, an annotation file must be loaded along with the microarray expression dataset to provide the gene names.
- Before overlaying the t-test results, any previously assigned colors will be cleared from the network display, regardless of their origin.
t-test example
Setup:
- This example begins with a network created in the Cellular_Networks_KnowledgeBase chapter. It is based on the BCell-100.exp dataset provided with geWorkbench, as well as the Affymetrix HG-U95Av2.na31.csv annotation file.
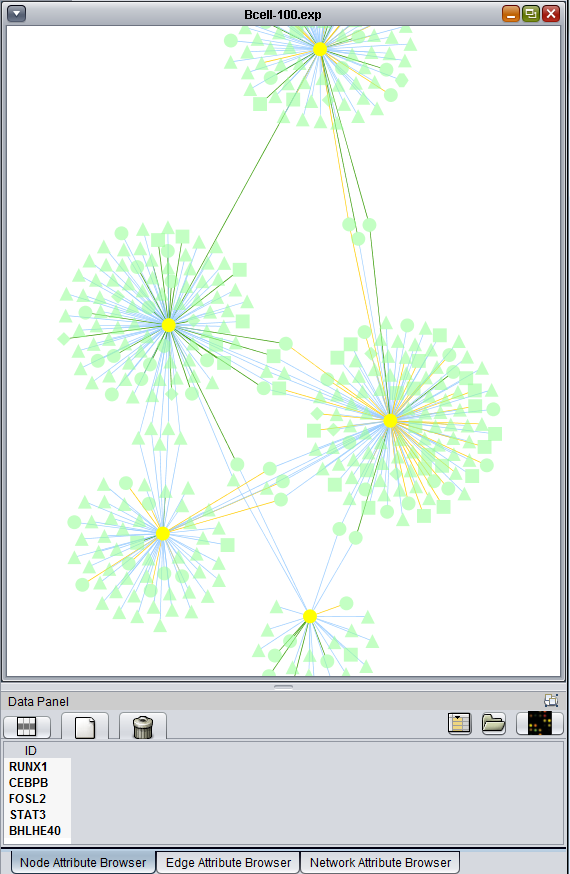
- A set of transcription factors was used to query the CNKB as described in the chapter Cellular_Networks_KnowledgeBase. The resulting network can be seen there and the query results are repeated below.
The data was prepared for the t-test by
- The BCell-100 data node was selected in the Workspace. (It should have a child adjacency matrix from the CNKB "create network" step).
- Normalization: threshold normalize to a minimum value of 1.0.
- Normalization: log2 transform.
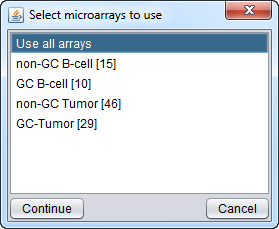
- Arrays Component: In the sets pulldown menu, select the array grouping "Class".
- Arrays Component: activate the sets "GC B-cell" and "GC tumor".
- Arrays Component: set the set "GC tumor" to be type "Case".
A t-test was computed on the BCell-100 dataset. The parameters used were
- p-value: 0.01
- Alpha Correction: Standard Bonferroni correction
- log2-transformed data box: checked.
After the t-test was calculated, the adjacency matrix from the CNKB query (with parent BCell-100) was selected in the Workspace to redisplay the network in Cytoscape.
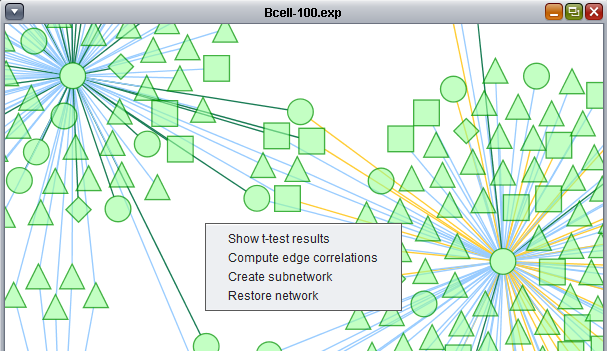
On the network display, the "Show t-test results" option was chosen.
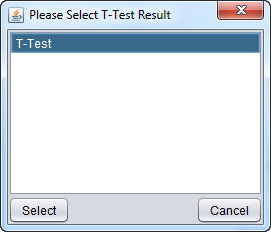
A dialog allows the t-test result to be displayed to be chosen, should there be more than one. Only t-test result nodes which belong to the same parent microarray expression dataset as the adjacency matrix being viewed in Cytoscape will be offered:
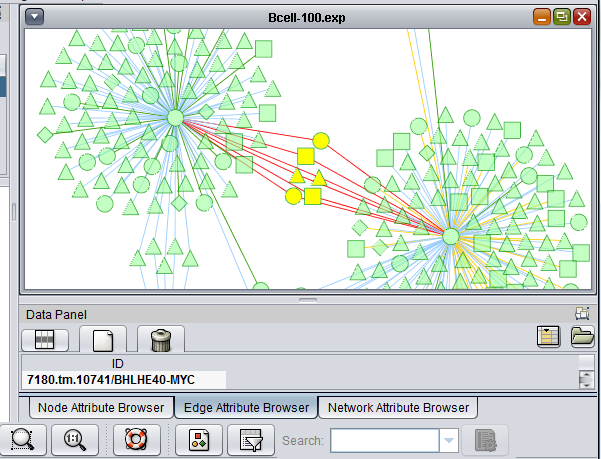
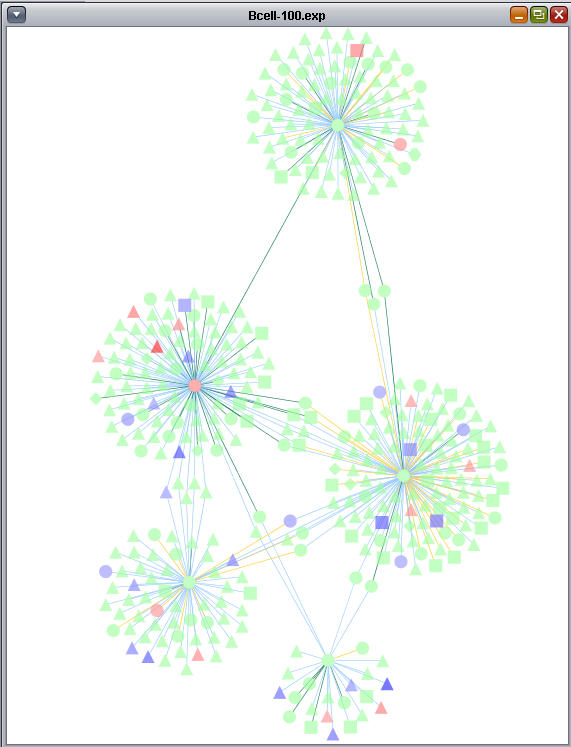
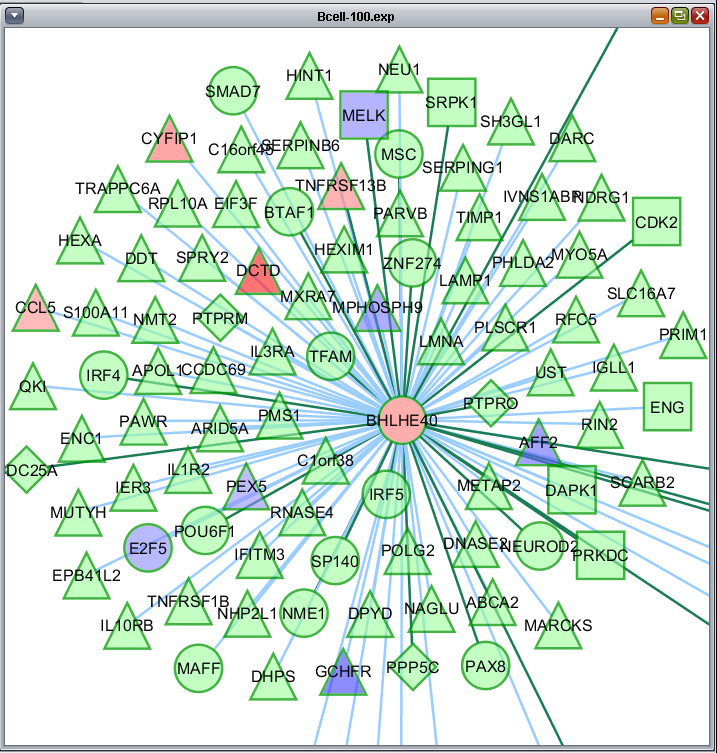
The t-test results are used to color the network nodes according to the sign and magnitude of their t-statistic. Positive values are shaded red, and negative values are shaded blue. The greater the magnitude of the value, positive or negative, the darker the shade of red or blue that is used.
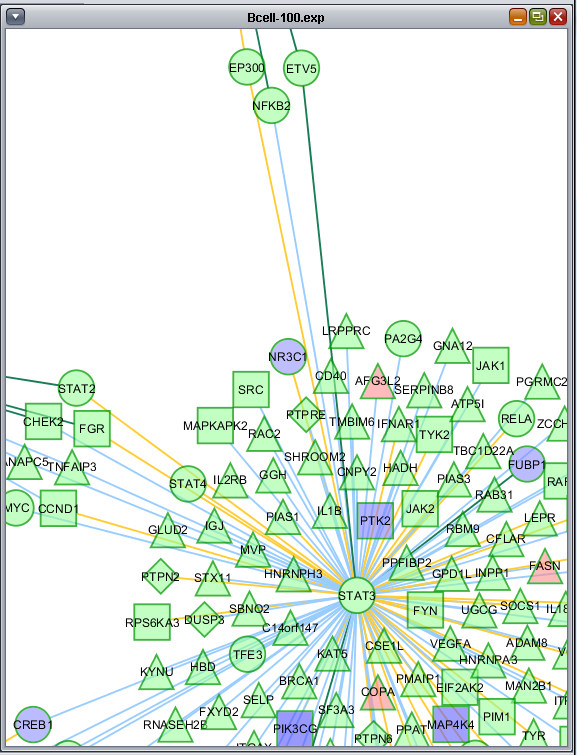
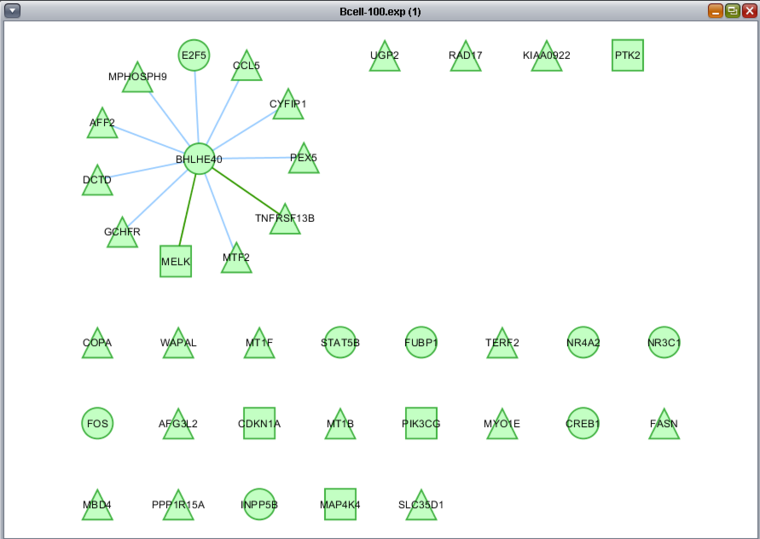
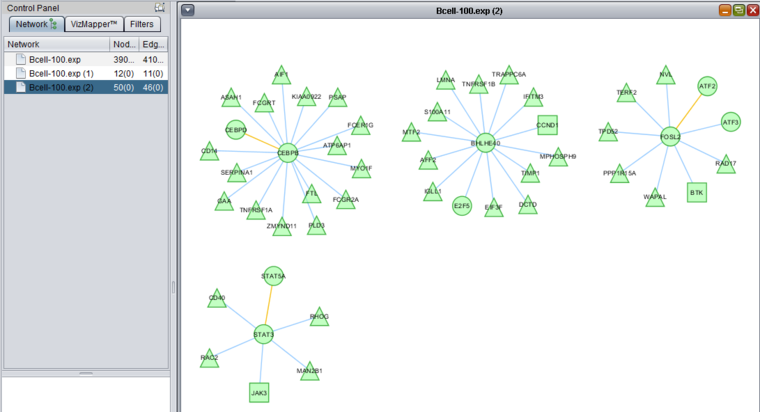
These two images show zoomed-in views of the network colored by t-test results.
Here is the network about BHLHE40, which is the one hub node that was colored in by the t-test results:
This figure shows all the different colorings of edges (based on interaction type) and nodes (based on t-test result).
Create subnetwork
The "Create subnetwork" feature intersects a network displayed in Cytoscape with a set of Markers defined in the Markers component.
- A new adjacency matrix is created in the Workspace. The new adjacency matrix is made a child of the same dataset as the network it was created from.
- The new subnetwork contains all nodes in the original network which are also in the selected marker set, regardless of whether they are connected by any edges.
Here we choose the "Significant Genes[428]" list of genes to intersect with the displayed network.
After the new subnetwork is created, it is placed as a new Adjacency matrix in the Workspace.
Restore network
Selecting "restore network" will remove any effects that have been applied to a particular adjacency matrix, such as overlaying t-test or Pearson's correlation results.
Specifically, it will
- restore the network to its original state,
- redraw all network nodes and edges,
- remove all node highlights,
- restore the original edge colors.
Note that creating a subnetwork actually creates a new adjacency matrix. If you wish to return to the original adjacency matrix, select it in the Workspace.
Compute Edge Correlations
Description
An network node (adjacency matrix) associated with a microarray gene expression dataset in geWorkbench may come from a source other than the microarray dataset itself. For example, it may have been loaded from disk, or fetched from the CNKB through a query. The user may wish to see how well this pre-computed network fits the expression data in the microarray set.
To that end, for each edge E in the network (that is, pairs of genes connected by an interaction), we can calculate its Pearson correlation r(E) from the data and then highlight only those edges for which |r(E)| is above a user-defined threshold (|r(E)| denotes the absolute value of the correlation r(E)).
For each pair of markers or genes connected by an edge, the Pearson's correlation of the expression profiles of those two genes in the microarray dataset is calculated using:
- if no annotation file has been loaded, or for other reasons gene names are not available, then all correlations will be computed at the marker level.
- if gene names are available, and if a gene is represented by more than one marker (probeset), the values for each such marker will be averaged by array to create one "average marker" for that gene. This "average marker" will then be used in the correlation calculation.
There are two possible ways in which the correlation value can be used:
- To create a new subnetwork (represented by a new adjacency matrix in the Workspace) which contains only those edges (and the nodes they connect) which met the correlation threshold, or
- To display on the original network only those edges which which met the correlation threshold. All nodes are still displayed. (In this case, the sub-threshold edges are invisible but are still present).
For purposes of this description, we continue working with the dataset described above in the t-test example.
When "Compute edge correlations" is chosen, a dialog appears in which the arrays to be included can be chosen.
The list of array sets shown in this dialog is the same as those currently visible in the Arrays component. In this case, these sets belong to the "Class" list, which was used to set up the t-test case and control groups.
In the figure below we select all arrays.
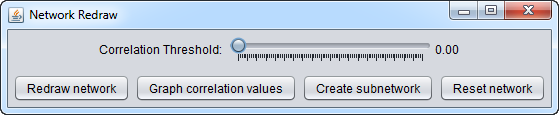
Next, one can choose among the several options:
- Graph correlation values
- Correlation threshold (slider control)
- Redraw network
- Create subnetwork
- Reset network
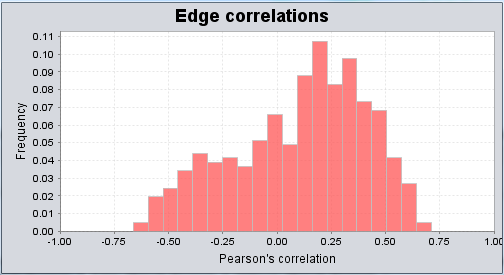
Graph Correlation Values
One can examine a histogram of the correlation values. This can aid in setting a reasonable cutoff for the correlation threshold used in visualizing the network overlap.
Correlation Threshold
This slider allows the minimum threshold for accepting an edge based on its Pearson's correlation coefficient to be set. Filtering-out of edges is based on the absolute value of the correlation coefficient.
In this example we set the slider to a cutoff value of 0.5.
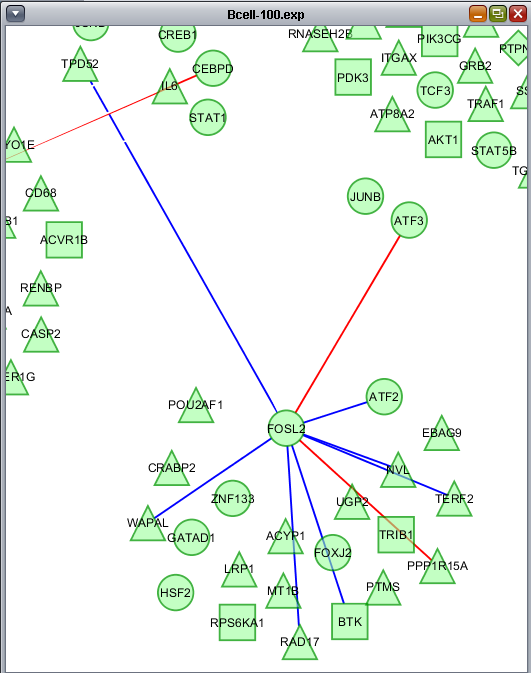
Redraw network
Only edges that meet the minimum threshold for the Pearson's correlation value are displayed, that is, only edges E for which |r(E)| >= T. All nodes are however retained in the underlying data. Edges are colored red or blue according to positive or negative correlation, respectively.
The below figure shows the retained edges in more detail.
Create subnetwork
Creates a subnetwork containing only nodes that are connected by above-threshold correlation results. Edges that do not meet the threshold, and also any unconnected nodes, are removed.
This step can either be performed directly after the threshold has been set, or after the "Redraw network" command has been used.
In either case, the subnetwork is represented by a newly created adjacency matrix in the Workspace. This new network node is created as a child of the same microarray dataset from which it was derived. Node attributes (like geneType) and edge attributes (like edge type) are copied over from the corresponding nodes and edges of the parent network.
To return to the original network, select its adjacency matrix in the Workspace.
(Note that this diagram was created with an edited set of interaction-type colors).
Reset network
The original network is redrawn, eliminating all correlation filtering of edges. Also, the original edge colors are restored.
References
Melissa S Cline, Michael Smoot, Ethan Cerami, Allan Kuchinsky, Nerius Landys, Chris Workman, Rowan Christmas, Iliana Avila-Campilo, Michael Creech, Benjamin Gross, Kristina Hanspers, Ruth Isserlin, Ryan Kelley, Sarah Killcoyne, Samad Lotia, Steven Maere, John Morris, Keiichiro Ono, Vuk Pavlovic, Alexander R Pico, Aditya Vailaya, Peng-Liang Wang, Annette Adler, Bruce R Conklin, Leroy Hood, Martin Kuiper, Chris Sander, Ilya Schmulevich, Benno Schwikowski, Guy J Warner, Trey Ideker & Gary D Bader. (2007) Integration of biological networks and gene expression data using Cytoscape. Nature Protocols 2, 2366 - 2382. Links: Article, PubMed
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Research 13(11):2498-504. Links: Article, PDF, PubMed.
The formal specification for the network edge filtering based on Pearson's correlation is found in Mantis issue #2424.