Local Data Files
Tutorial: Loading microarray data files - local
In this example, we will load 10 individual Affymetrix MAS5 format files, and merge them into a single dataset. The origin of these file is described in the section Tutorial_-_Data
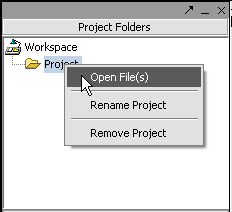
Next, right-click on the New Project entry and select Open Files.
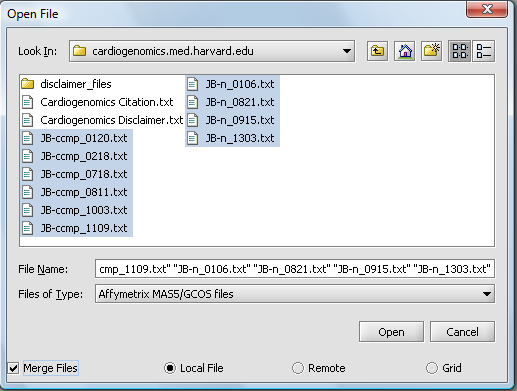
A file browser will appear with which you can select the files you wish to open. The default is to browse for local files, that is on your own computer. geWorkbench can also access data from caArray databases.
Here, we will select file type Affymetrix GCOS/MAS5 as shown.
Make sure to check the Merge files checkbox. This will create the merged data node as the files are read in.
We will select 10 MAS5 format text files from the directory cardiogenomics.med.harvard.edu, which is included in the geWorkbench tutorial download.
Click Open.
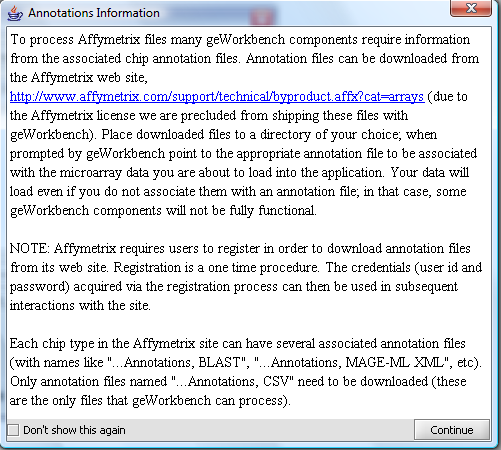
A message will appear giving information about associating an annotation file with the dataset.
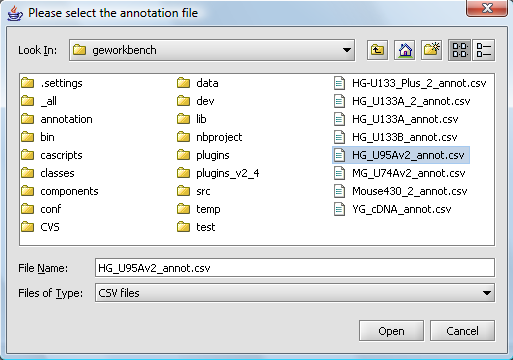
A file browser will then open with which you can, if you wish, select an annotation file matching your dataset. This is needed if you intend to use features of geWorkbench such as the Sequence Retriever or GO Terms component (Gene Ontology).
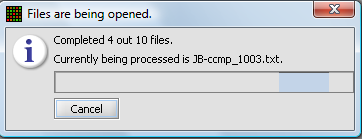
A status bar will display as the data is loaded:
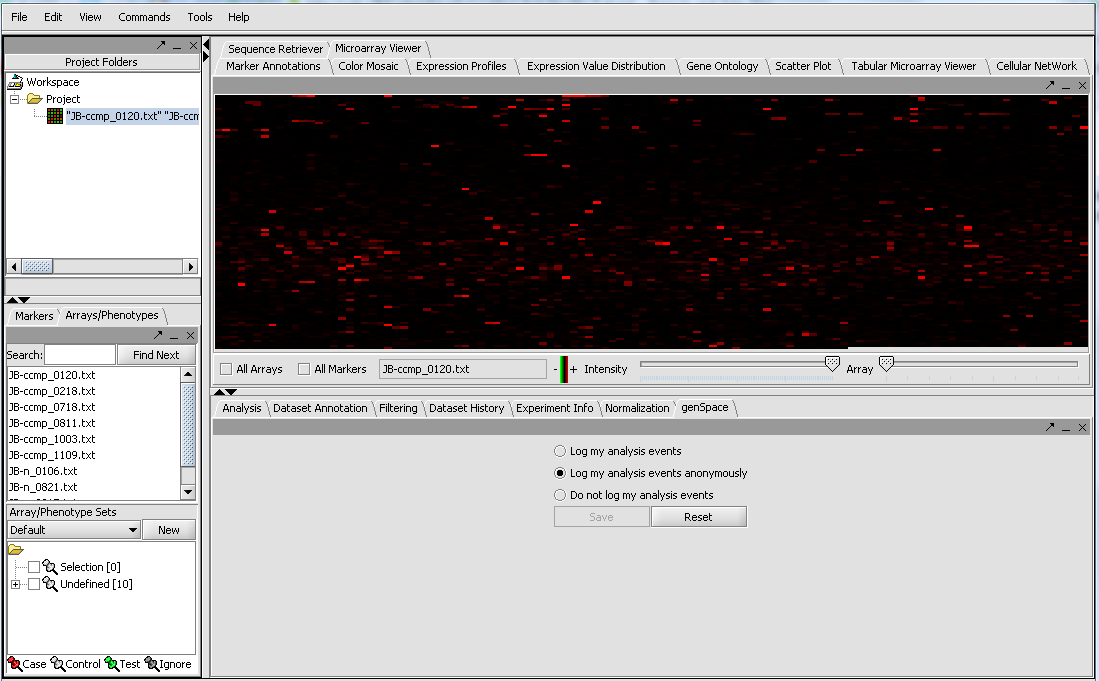
The merged dataset is listed in the Project folder. The data is displayed, in single array format, in the Microarray Viewer. Note we have increased the intensity slider to maximum here. You can scroll through the arrays from first to last using the slider. The display in the Microarray Viewer is by marker in the linear order the markers appear in the data file. It does not correspond in any way to a physical picture or representation of the actual 2-D microarray.
Tutorial: Merging microarray data files after they have already been loaded.
If data files are not merged at the time they are read in, they can also be merged later, as long as they are from the same chip type.
1. Select the read-in data files that you want to merge.
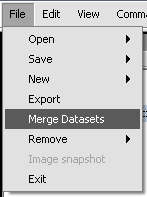
2. Click on File in the menu bar, and choose Merge Datasets.
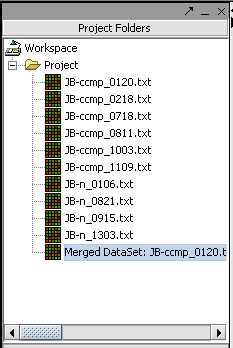
The picture shows the resulting merged dataset created from several individual data files.
The result is a new data node containing the merged data. The original data nodes are still present.