Difference between revisions of "Normalization"
m (→Quantile Normalization) |
(→Housekeeping Gene Normalization) |
||
| Line 71: | Line 71: | ||
[[Image:Normalization-HouseKeeping.png]] | [[Image:Normalization-HouseKeeping.png]] | ||
| + | This component allows one to load a list of housekeeping gene markers ('''Load''' button). Loaded markers initially appear in the list at right, "Current Selected Genes". Markers can be moved back and forth between this list and the list at left, "Excluded Genes" through use of the right and left arrow buttons ('''< >'''), or simply by double-clicking on a marker in either list. | ||
| + | |||
| + | '''Right-click menu''' - If one right-clicks on a marker in the "Current Selected Genes" list, a pop-up menu appears, offering two choices: | ||
| + | # '''Rename''' - will rename the marker in the list. Note that the marker name must match a marker in the currently loaded dataset. | ||
| + | # '''Delete''' - delete a marker from the list. | ||
| + | |||
| + | ===Controls=== | ||
| + | * '''<, >''' = Shift selected markers between lists. | ||
| + | * '''Clear All''' - clear both lists. | ||
| + | * '''Load''' - load a list of markers from a comma-separated value (CSV) format file. One marker per line is also an acceptable format. | ||
| + | * '''Save''' - Save the list shown in the "Current Selected Genes" list to a file. The output file format is one marker per line. | ||
| + | |||
| + | ===Missing Value Options for Housekeeping Genes=== | ||
| + | * '''Ignore''' - do not replace missing values. | ||
| + | * '''Microarray Average''' - missing values are replaced for the housekeeping gene normalization with the average of all values on the microarray on which the missing value was found. | ||
==Log2 Transformation== | ==Log2 Transformation== | ||
Revision as of 11:53, 8 June 2010
Contents
- 1 Overview
- 2 Normalizer Configuration
- 3 Available Normalizers
- 4 Basic Controls
- 5 Specific Controls for each Normalizer
Overview
Normalization is used to transform data in preparation for analysis. Many normalizers are oriented towards decreasing the effects of systematic differences across a set of microarrays, aiding in cross-microarray comparisons. A log transformation of the data can be used to improve its statistical distribution. geWorkbench offers a selection of pluggable normalization components (see list below). The Normalizer panel is located in the Commands/ Analysis area in the lower right side of the application.</p>
In geWorkbench, normalization alters the loaded dataset and is not reversible; the original is not retained. Normalization operations do not respect any marker or array sets that may be activated; normalization always acts on the entire data set.
The Missing Value Computation normalizer can be used to replace missing values with imputed values. Note that some analysis routines also will optionally compute replacements for missing values if needed.
Data preparation methods such as RMA and GCRMA are not directly available in geWorkbench. Affymetrix CEL files can be processed externally to geWorkbench using a program such as RMAExpress or in R/Bioconductor and then the processed data imported into geWorkbench.
A short introduction to various methods of Affymetrix data preparation is available at Affymetrix Preprocessing.
Normalizer Configuration
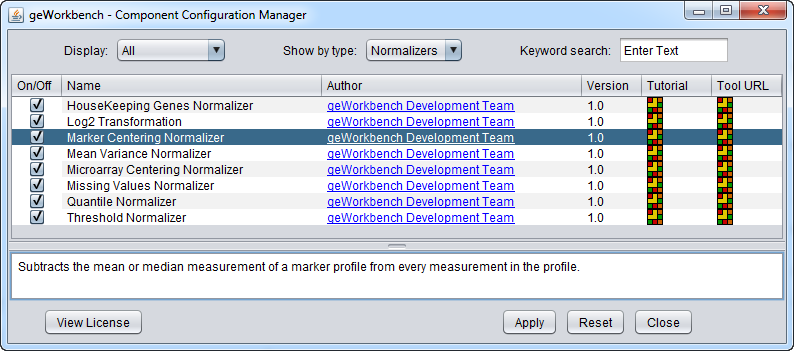
Some normalizers may not be loaded by default in geWorkbench. To configure which normalizers to load, use the Component Configuration Manager. It is available in the top menu-bar under Tools->Component Configuration.
Available Normalizers
geWorkbench comes with the following normalization routines installed:
| Normalizer | Description | |
|---|---|---|
| Housekeeping Genes Normalization | Normalize all values such that the averaged expression value of specified house-keeping markers is the same on each microarray. | |
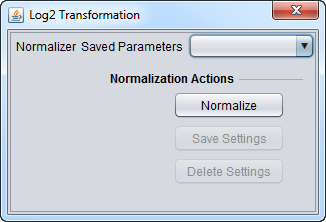
| Log2 Transformation | Applies a log2 transformation to all measurements in a microarray. | |
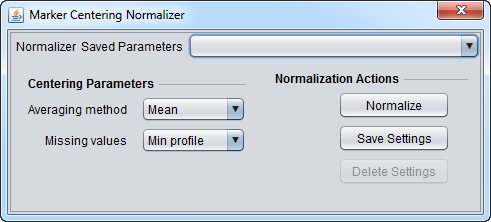
| Marker-based Centering | Subtracts the mean or median measurement of a marker profile from every measurement in the profile. | |
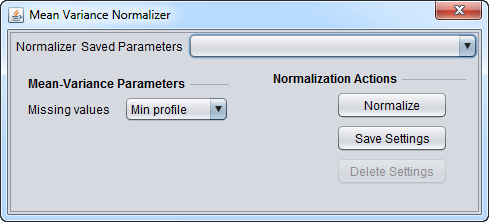
| Mean-variance Normalization | For every marker profile, the mean measurement of the entire profile is subtracted from each measurement in the profile and the resulting value is divided by the standard deviation. | |
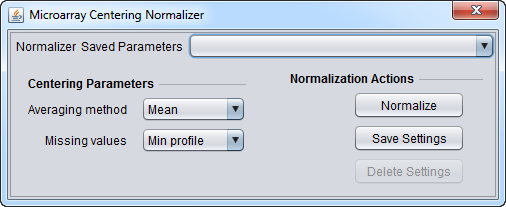
| Microarray-based Centering | Subtracts the mean or median measurement of a microarray from every measurement in that microarray. | |
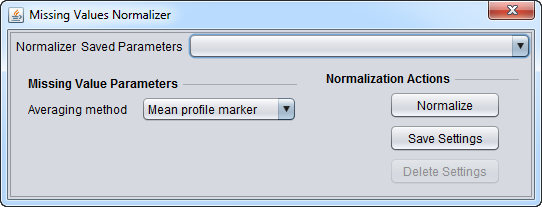
| Missing Value Computation | Replaces every missing value with either the mean value of that marker across all microarrays or with the mean measurement of all markers in the microarray where the missing value is observed. | |
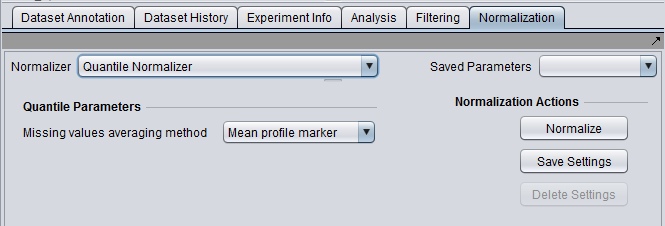
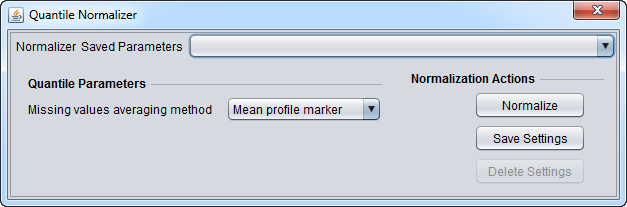
| Quantile Normalization | Adjusts expression values so that the distribution of values is the same on each microarray, though which marker has which value varies. | |
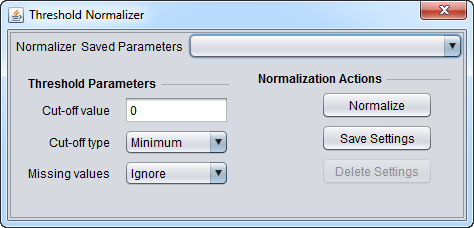
| Threshold Normalization | All data points whose value is less than (or greater than) a user-specified minimum (maximum) value are raised (reduced) to that minimum (maximum) value. |
Basic Controls
Overview
- Normalizer (menu) - select the desired normalizer.
- Save Parameters (menu) - allows selection of stored parameter settings.
- Normalize (button)- run the selected transformation.
- Save Settings - Save the current settings (see following section).
- Delete Settings- delete the currently selected parameter set.
Saving Parameters
The current parameter settings can be saved to a named parameter set. The saved set will be displayed in the pull-down menu at upper right in the component. Any number of parameter sets can be saved. If the currently set parameters match a saved set, that set's entry will be shown in the menu.
- Save Settings - save the current settings to a new parameter set.
- Delete Settings - delete the currently selected parameter set from the menu.
Specific Controls for each Normalizer
Housekeeping Gene Normalization
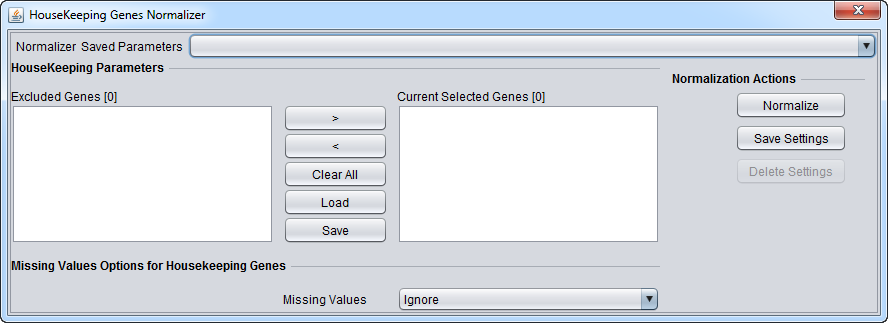
This component allows one to load a list of housekeeping gene markers (Load button). Loaded markers initially appear in the list at right, "Current Selected Genes". Markers can be moved back and forth between this list and the list at left, "Excluded Genes" through use of the right and left arrow buttons (< >), or simply by double-clicking on a marker in either list.
Right-click menu - If one right-clicks on a marker in the "Current Selected Genes" list, a pop-up menu appears, offering two choices:
- Rename - will rename the marker in the list. Note that the marker name must match a marker in the currently loaded dataset.
- Delete - delete a marker from the list.
Controls
- <, > = Shift selected markers between lists.
- Clear All - clear both lists.
- Load - load a list of markers from a comma-separated value (CSV) format file. One marker per line is also an acceptable format.
- Save - Save the list shown in the "Current Selected Genes" list to a file. The output file format is one marker per line.
Missing Value Options for Housekeeping Genes
- Ignore - do not replace missing values.
- Microarray Average - missing values are replaced for the housekeeping gene normalization with the average of all values on the microarray on which the missing value was found.