Difference between revisions of "Viper Analysis"
(→Parameters) |
|||
| Line 16: | Line 16: | ||
* '''Select Method''' | * '''Select Method''' | ||
| − | ** '''none''' - | + | ** '''none''' - no correction. |
** '''scale''' - for each gene (row in dataset), calculate the mean and standard deviation across all columns, then subtract the mean from each value in the row, and divide each by the standard deviation. | ** '''scale''' - for each gene (row in dataset), calculate the mean and standard deviation across all columns, then subtract the mean from each value in the row, and divide each by the standard deviation. | ||
** '''rank''' - | ** '''rank''' - | ||
** '''mad''' - | ** '''mad''' - | ||
** ''ttest''' - | ** ''ttest''' - | ||
Revision as of 14:09, 21 November 2013
Page in preparation, should be available 11/22/2013.
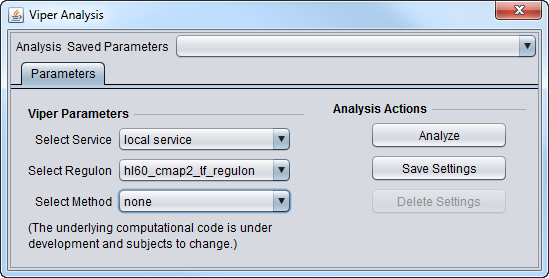
Parameters
- Select Service
- Local Service - run VIPER on an instance of R installed on the same machine as geWorkbench.
- Web service - not yet implemented. Run Viper on a remote server.
- Select Regulon
- hl60_cmap2_tf_regulon - B-cell lymphoma, CMAP2 data
- mcf7_cmap2_tf_regulon - Breast cancer, CMAP2 data
- pc3_cmap2_tf_regulon - Prostate cancer, CMAP2 data
- Select Method
- none - no correction.
- scale - for each gene (row in dataset), calculate the mean and standard deviation across all columns, then subtract the mean from each value in the row, and divide each by the standard deviation.
- rank -
- mad -
- ttest' -