Viper Analysis
Page in preparation, should be available 11/22/2013.
(Virtual Inference of Protein-activity by Enriched Regulon analysis)
The VIPER component in geWorkbench transforms the expression profile for each sample (column) into a transcription-factor activity profile. The activity of each transcription factor is inferred from that of its targets, where the targets are obtained from a cell-context-specific interaction network (interactome). Three cell-context-specific interactomes are supplied, for leukemia, breast cancer, and prostate cancer.
The full standalone version of VIPER can also be downloaded from http://wiki.c2b2.columbia.edu/califanolab/index.php/Software/VIPER. VIPER is implemented in R and the standalone package has a number of additional functions.
In geWorkbench, the most simple variant of VIPER is employed, which assumes that in the null situation the target genes are uniformly distributed on the gene expression signature. The standalone version offers a permutation method (given a set of control samples) to calculate a null model accounting for non-independence of expression between genes.
VIPER and its source code are released in geWorkbench under the VIPER Software License.
Contents
Data
VIPER requires expression data that is already in a form where the values for each sample are relative to a control value. Each sample may represent e.g. the result of a drug perturbation.
Analysis
For a typical dataset containing expression values relative to control, the "Scale" method of analysis is recommended.
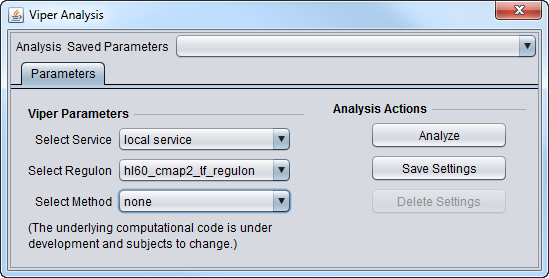
Parameters
- Select Service
- Local Service - run VIPER on an instance of R installed on the same machine as geWorkbench.
- Web service - not yet implemented. Run Viper on a remote server.
- Select Regulon
- hl60_cmap2_tf_regulon - Human promyelocytic leukemia, CMAP2 data
- mcf7_cmap2_tf_regulon - Breast adenocarcinoma, CMAP2 data
- pc3_cmap2_tf_regulon - Prostate cancer, CMAP2 data
- Select Method
- none - use if the data is already in rank format.
- scale - for each gene (row in dataset), calculate the mean and standard deviation across all columns, then subtract the mean from each value in the row, and divide each by the standard deviation.
- rank - rank transform row-wise
- mad - subtract the median, divide by the mean absolute deviation (MAD)
- ttest - for each gene, perform a t-test comparing each sample (column) one-at-a-time against all other samples taken together.
References
Alvarez et al, manuscript in preparation.