User:Ginhoven
TUTORIAL - BLAST
In this Tutorial you will learn to:
- Set up and perform a Blast search.
- Decipher the Output.
- Analyze the results.
OVERVIEW
A reason why you may want to do a BLAST Query may be that you have found an interesting marker, so you want to retrieve it's gene, and see what it is related to.
BLAST searches are divided into categories according to the nature, and size of the input query and the primary goal of the search.
A BLAST search has four components:
- Query
- Data Base Program
- Search Purpose
- Goal
For the purpose of this tutorial use the file "NM _024426-Wilms.Fasta" provided in the tutorial data directory. This is a nucleotide sequence file. There is a second file which contains the corresponding protein sequence "NP_077744-Wilms.fasta".
Provide a little background info about Wilm's tumor. (It was chosen at random).
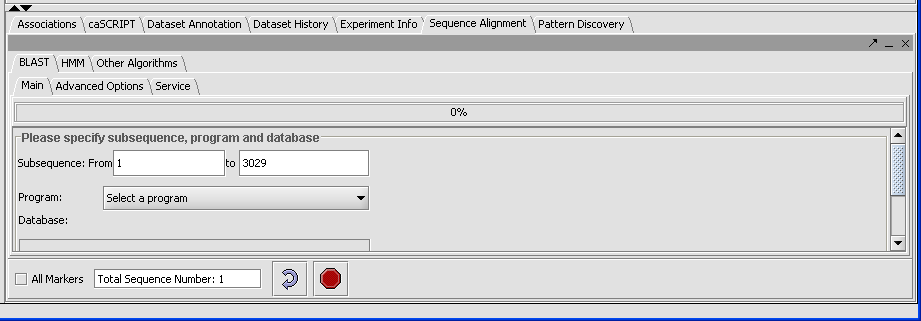
- In the Visualization Area click on the Sequence Alignment tab.
- Click on the Blast tab.
The result displays the length of the longest sequence selected (here there is only one) due to the sample.
There are five different types of queries you can run, depending on what data you are using:
blastp- Compares an amino acid query sequence against a protein sequence database.
blastn- Compares a nucleotide query sequence against a nucleotide sequence database.
blastx- Compares a nucleotide query sequence translated in all reading frames against a protein sequence database.
tblastn- Compares a protein query sequence against a nucleotide database dynamically translated in all reading frames.
tblastx- Compares the 6 frame translations of a nucleotide query sequence against the six frame translations of a nucleotide sequence database.
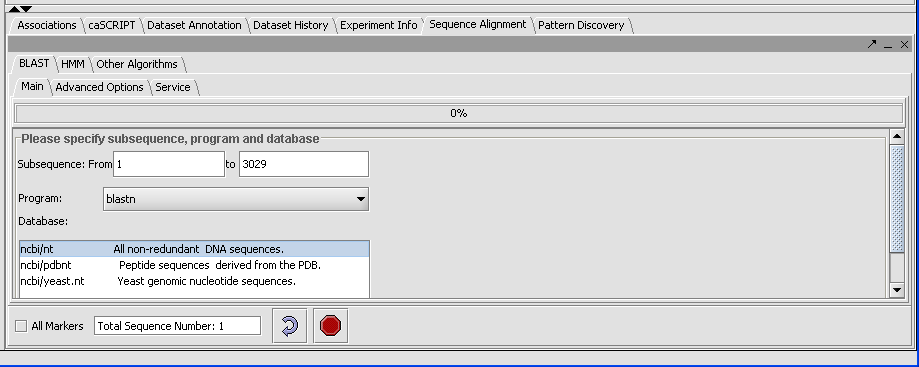
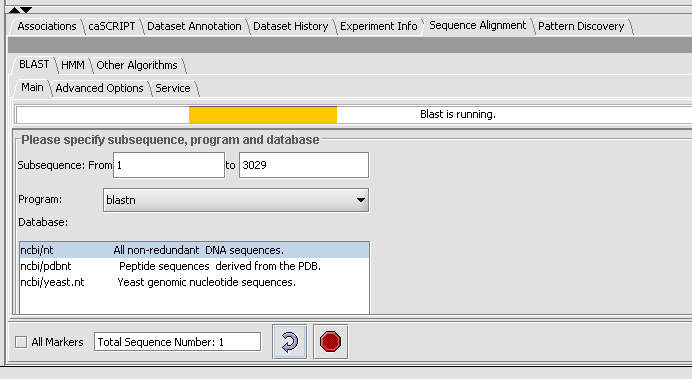
Click on the drop down arrow and select a program. Since this is a nucelotide query, we want to select a nucleotide query program blastn.
Now that the program has been selected, make sure the appropriate databases are displayed (you need to verify this for all algorithms). Here select ncbi/nt - the complete non-redundant nucleotide database.
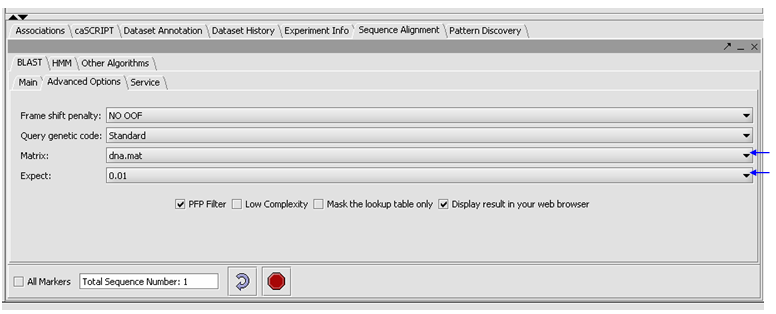
- Click on the Advanced Options Tab
- Make sure "dna mat" is selected for the Matrix.
- Change the Expect Value to 0.01.
- Leave the box checked for PFP filtering for repeated sequence elements (Paracel Filtering Package).
- Leave the Display result in your web browser checked.
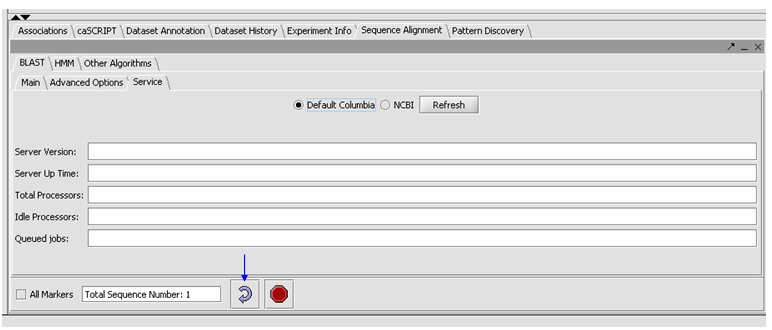
- Click on the Service tab, select Columbia.
Note: The text field at the bottom shows that one sequence has been selected. If you have a Fasta file that has multiple sequences, you can select the ones you want in the Markers component and activate this selection, letting you search on a subset. You may search on all sequences in a file by clicking the All Markers checkbox.
- Press the curved arrow submit button.
- Observe the progress bar, Blast is now runnning.
- You can check the server status by hitting the Refresh button, under the Service tab. Then you will an idea if the Columbia Machine is processing a lot of queries.
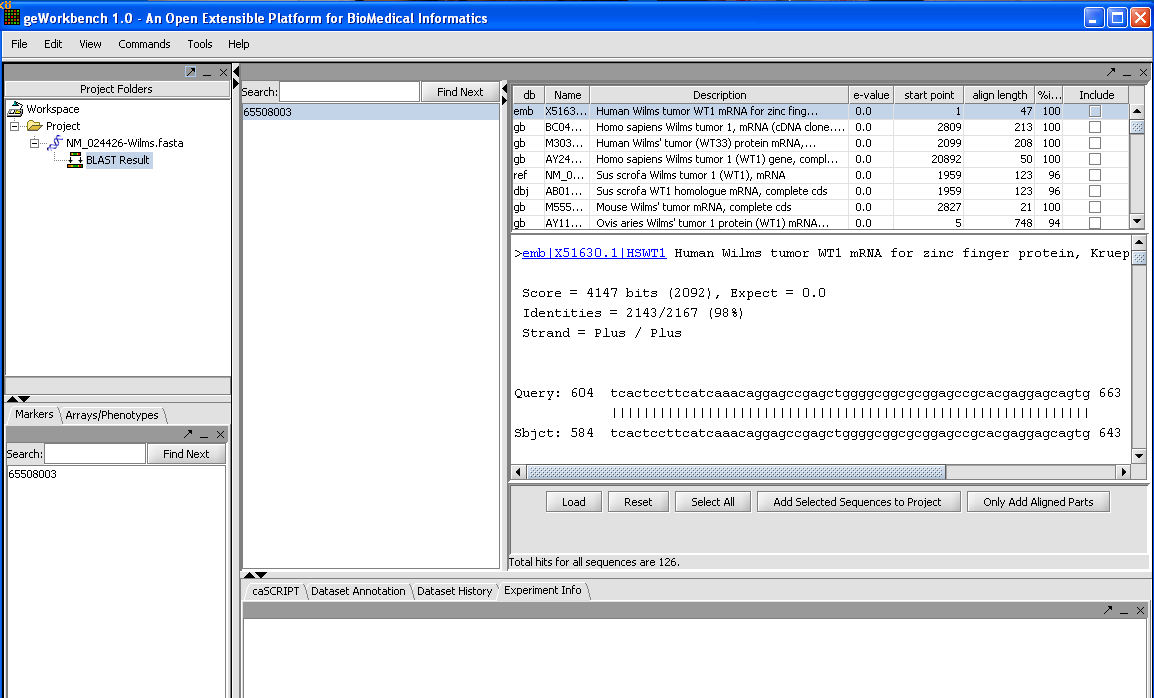
When the results are returned they are placed in the Project Folders as a child of the sequence they correspond to. You can mouse over the result set to see how many sequences are in it.
In the Blast results viewer, you can examine the alignments. Each different target hit is listed on a line in the results table.
In the Blast results viewer you can select sequences to add back to the main project by checking the include box and then the Add Selected Sequences To Your Project tab.
You can also add just the aligned parts by clicking on the tab Only Add Aligned Parts.
(Should we say anything about e value and bit score in this tutorial?) Do we need more details on reading the data output? We need to explain more about the output. What about the separate page that pops up?
The Load button allows you to load an external Blast file in HTML format into the viewer.