Difference between revisions of "MRA-FET"
| Line 18: | Line 18: | ||
* '''Plus mode''' - the expression profile of the TF is positively correlated with those of regulon markers showing positive differential expression in the "case" set. The TF is more active in the "case" state. | * '''Plus mode''' - the expression profile of the TF is positively correlated with those of regulon markers showing positive differential expression in the "case" set. The TF is more active in the "case" state. | ||
* '''Minus mode''' - the expression profile of the TF is positively correlated with those of regulon markers showing negative differential expression in the "case" set. The TF is more active in the "control" state. | * '''Minus mode''' - the expression profile of the TF is positively correlated with those of regulon markers showing negative differential expression in the "case" set. The TF is more active in the "control" state. | ||
| + | |||
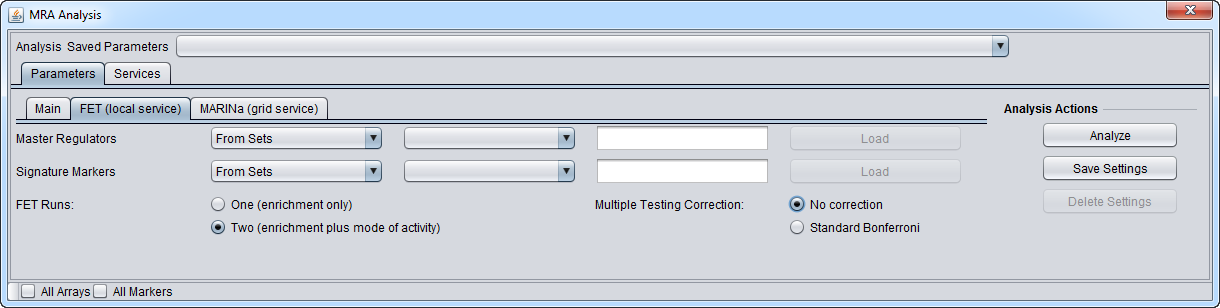
| + | =MRA-FET (local service)Parameters= | ||
| + | |||
| + | [[Image:MRA_Parameters_FET_v2.png]] | ||
| + | |||
| + | ==Master Regulators== | ||
| + | A set of candidate master regulator markers. | ||
| + | * This set must be loaded into the Markers component before running MRA. The set can be created directly there, or read in from a file. | ||
| + | |||
| + | ==Signature Markers== | ||
| + | A set of markers comprising the signature that distinguishes the chosen phenotype from others. | ||
| + | * This set must be loaded into the Markers component before running MRA. The signature can be generated directly, e.g. through a t-test, or loaded from a file. | ||
| + | |||
| + | ==FET Runs== | ||
| + | * One (Enrichment Only) | ||
| + | * Two (Enrichment plus mode of activity) - the target markers are divided into two groups and two runs of FET are performed. See the description above at [[Master_Regulator_Analysis#FET_method_details | FET method details]]. | ||
| + | |||
| + | ==Multiple Testing Correction== | ||
| + | * No Correction | ||
| + | * Standard Bonferroni | ||
| + | |||
| + | |||
| + | ==T-test for differential expression== | ||
| + | In the Arrays component, a case and a control group must be defined for running a t-test. | ||
| + | |||
| + | A "bar-code" graphic is generated using a t-test of differential expression. However, all t-values are accepted (critical alpha = 1) and used to order the bars representing the regulon markers. | ||
| + | |||
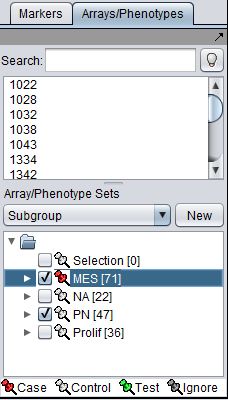
| + | All that is required is to define sets of arrays representing two phenotypes of interest (and distinguished by the signature). At least two sets of arrays must be activated, and at least one marked as "case", representing the target phenotype of the gene signature. "Control" is the default classification. See also the [[Differential_Expression | Differential Expression tutorial]]). | ||
| + | |||
| + | |||
| + | [[Image:Array_set_class_assignment_MRA.png]] | ||
Revision as of 14:29, 26 November 2012
Contents
FET method details
Description
Two choices are available in how to apply the FET method.
- One FET run - a single run of FET is used to determine enrichment of the signature markers in the hub's regulon.
- Two FET runs - The data is divided into two subsets based on
- positive or negative differential expression of targets, and also into two orthogonal subsets based on
- positive or negative Spearman's Correlation of the expression of the targets across all arrays (not just those used in the test of differential expression) as compared to the hub markers.
TF Activity Modes
Using the notation (differential expression result, Spearman's correlation result) for the intersection of differential expression (+ or -) and correlation (+ or -) results, the following two sets are formed and FET is run for each:
- Test 1 (plus mode): (+,+) union (-,-).
- Test 2 (minus mode): (+,-) union (-,+).
Whichever of the two tests gives the more significant p-value is used as the final p-value and the mode is called as "plus" or "minus" correspondingly. The mode is displayed in the MRA results viewer.
Simplified Interpretation of Modes
- Plus mode - the expression profile of the TF is positively correlated with those of regulon markers showing positive differential expression in the "case" set. The TF is more active in the "case" state.
- Minus mode - the expression profile of the TF is positively correlated with those of regulon markers showing negative differential expression in the "case" set. The TF is more active in the "control" state.
MRA-FET (local service)Parameters
Master Regulators
A set of candidate master regulator markers.
- This set must be loaded into the Markers component before running MRA. The set can be created directly there, or read in from a file.
Signature Markers
A set of markers comprising the signature that distinguishes the chosen phenotype from others.
- This set must be loaded into the Markers component before running MRA. The signature can be generated directly, e.g. through a t-test, or loaded from a file.
FET Runs
- One (Enrichment Only)
- Two (Enrichment plus mode of activity) - the target markers are divided into two groups and two runs of FET are performed. See the description above at FET method details.
Multiple Testing Correction
- No Correction
- Standard Bonferroni
T-test for differential expression
In the Arrays component, a case and a control group must be defined for running a t-test.
A "bar-code" graphic is generated using a t-test of differential expression. However, all t-values are accepted (critical alpha = 1) and used to order the bars representing the regulon markers.
All that is required is to define sets of arrays representing two phenotypes of interest (and distinguished by the signature). At least two sets of arrays must be activated, and at least one marked as "case", representing the target phenotype of the gene signature. "Control" is the default classification. See also the Differential Expression tutorial).