Classification
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Outline
This tutorial contains
- an overview describing two classification algorithms which run on a GenePattern server, and that are available through geWorkbench: (i) K-Nearest Neighbors (KNN), and (ii) Weighted Voting,
- a detailed example of setting up and running a KNN classification,
- an similar example of running the Weighted Voting classification.
A third method, SVN is described on the separate SVN tutorial page.
Overview
Classification algorithms are used to assign new experimental data sets to particular types based on prior training with known datasets. For example, they could be used to distinguish whether a new tissue sample was from a cancerous or normal tissue. There are many such classifiers available and they differ in which kind of data they are most appropriate for.
At present, geWorkbench provides interfaces to three GenePattern classification algorithms available on GenePattern servers:
- WV Classifier (Weighted-Voting)
- KNN Classifier (K-Nearest Neighbors)
- SVM - see the separate SVM tutorial page.
KNN Classifier - K-Nearest Neighbor
From the GenePattern KNN module documentation:
The K-Nearest Neighbor algorithm classifies a sample by assigning it the label most frequently represented among the k nearest samples. No explicit model for the probability density of the classes is formed; each point is estimated locally from the surrounding points. Target classes for prediction (classes 0 and 1) can be defined based on a phenotype such as morphological class or treatment outcome. The class predictor is uniquely defined by the initial set of samples and marker genes. The K-Nearest Neighbor algorithm stores the training instances and uses a distance function to determine which k members of the training set are closest to an unknown test instance. Once the k-nearest training instances have been found, their class assignments are used to predict the class for the test instance by a majority vote. Our implementation of the K-Nearest Neighbor algorithm allows the votes of the k neighbors to be un-weighted, weighted by the reciprocal of the rank of the neighbor's distance (e.g., the closest neighbor is given weight 1/1, next closest neighbor is given weight 1/2, etc.), or by the reciprocal of the distance. Either the Cosine or Euclidean distance measures can be used. The confidence is the proportion of votes for the winning class. There are many references for this type of classifier (with several of the early important papers listed below).
WV Classifier - weighted voting
From the GenePattern WV module documentation:
The Weighted Voting algorithm makes a weighted linear combination of relevant “marker” or “informative” features obtained in the training set to provide a classification scheme for new samples. Target classes (classes 0 and 1) can be for example defined based on a phenotype such as morphological class or treatment outcome. The selection of classifier input features (marker features) is accomplished either by computing a signal-to-noise statistic Sx = (μ0 - μ1)/( σ0 + σ1) where μ0 is the mean of class 0 and σ0 is the standard deviation of class 0 or by reading in a list of user provided features. The class predictor is uniquely defined by the initial set of samples and markers. In addition to computing Sx, the algorithm also finds the decision boundaries (half way) between the class means: Bx = (μ0 + μ1)/2 for each feature x. To predict the class of a test sample y, each feature x in the feature set casts a vote: Vx = Sx (Gxy – Bx) and the final vote for class 0 or 1 is sign(Sx Vx). The strength or confidence in the prediction of the winning class is (Vwin - Vlose)/(Vwin + Vlose) (i.e., the relative margin of victory for the vote). Notice that this algorithm is quite similar to Naïve Bayes (see the appendix in Slonim et al. 2000).
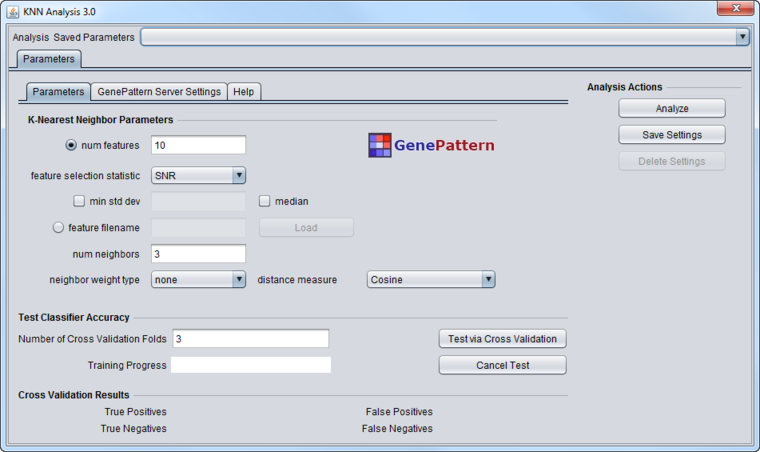
Graphical User Interface
The GUI is divided into two sections. The upper section allows parameters to be set for the actual classification run. The lower section allows parameters to be set for generation and cross-validation of the classifiers.
KNN Classifier Parameters
- num features - number of signal-to-noise selected features if "feature filename" is not specified
- feature selection statistic - statistic to use to perform feature selection if "feature filename" is not specified. Possible values are:
- SNR - Signal to Noise Ratio
- T-test
- min std dev - minimum standard deviation to be used by "feature selection statistic" (optional)
- median - whether median should be used by "feature selection statistic"
- feature filename - list of features to use for prediction (overrides feature selection parameters)
- num neighbors - number of neighbors for KNN
- neighbor weight type - weighting type of neighbors for KNN
- none
- 1/k
- distance
- distance measure - distance measure for KNN
- Cosine
- Euclidean
WV Classifier Parameters
- num features - number of signal-to-noise selected features if "feature filename" is not specified
- feature selection statistic - statistic to use to perform feature selection if "feature filename" is not specified. Possible values are:
- SNR - Signal to Noise Ratio
- T-test
- min std dev - minimum standard deviation to be used by "feature selection statistic" (optional)
- median - whether median should be used by "feature selection statistic" (optional)
- feature filename - list of features to use for prediction (overrides feature selection parameters)
Test Classifier Accuracy
- Number of Cross-Validation Folds - how many runs of cross-validation will be done. Must be greater than 1.
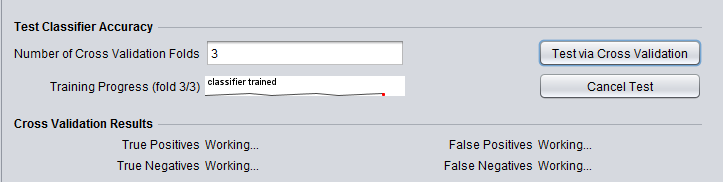
- Training Progress - a graphical depiction of percent job completion.
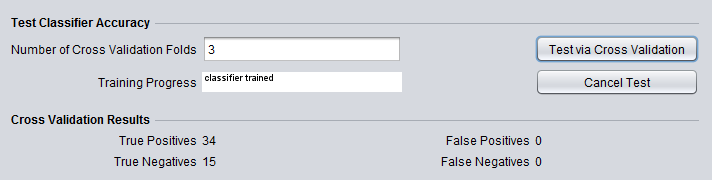
- Cross Validation Results - Shows true and false positives and negatives from the cross-validation step.
Example - K-Nearest Neighbors (KNN)
Prerequisites
A microarray data set must be loaded. For this example we will use the Bcell-100.zip (5.256 MB) dataset, also included in the geWorkbench "data/public_data" folder.
Setting up the Arrays
For the classification run, three sets of data must be provided - case, control, and a dataset on which the resulting classifier will be tested.
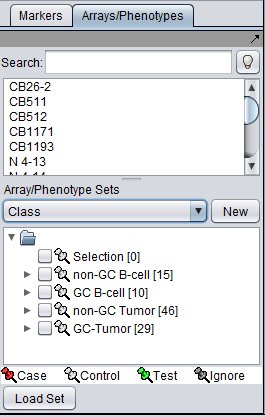
The figure below shows the four broad classes present in the Bcell-100.exp tutorial dataset. Note that we are viewing the Array/Phenotype list named "Class". We will use the non-GC B-cell and non-GC tumor classes.
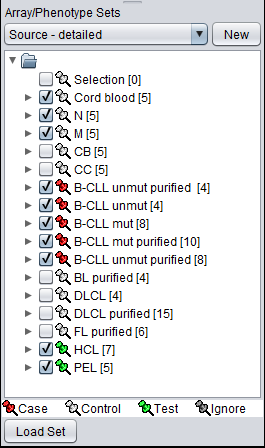
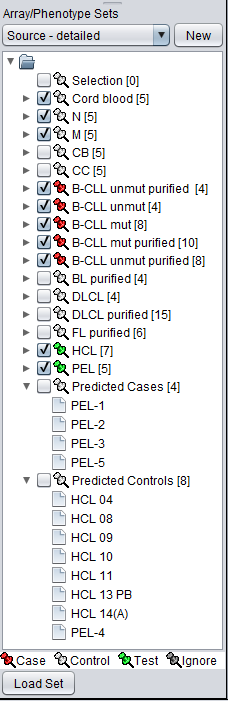
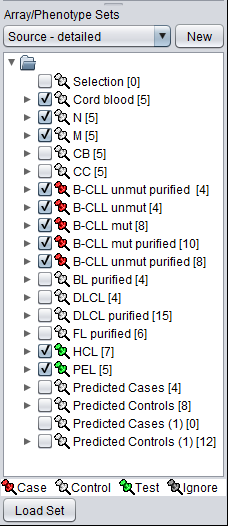
If we now change to the array list named "Source - detailed", we see a breakdown of the samples into more detailed cell line designations. By comparing with the members in "Class", we see we can set up the test as follows:
- Control: non-GC (non-GC B-cell)
- Case: B-CLL (non-GC tumor)
- Test: HCL, PEL (non-GC tumor)
Thus the expected outcome of the classification would be that all members of the test set would be identified as members of the Case group.
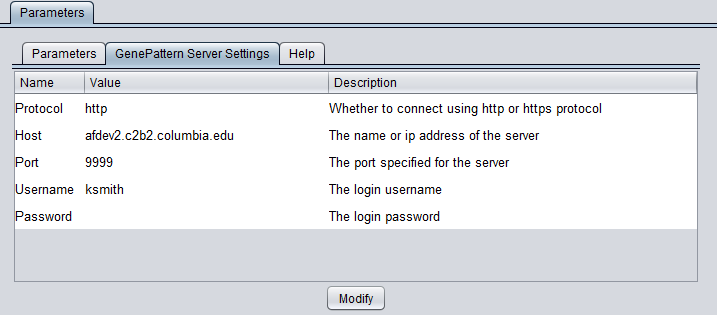
GenePattern Server Settings
You can connect to any running GenePattern server to run the analysis (provided it has the required module installed). An example configuration of the "GenePattern Server Settings" tab is shown here:
To run GenePattern components, a GenePattern account is required.
Pushing "Modify" brings up an editing box where any of the settings can be changed.
- Protocol - HTTP or HTTPS, depending on the server being used.
- Host - URL of a GenePattern server.
- Port - Port at which the GenePattern server is located on the Host machine.
- Username - A valid user name on the specified GenePattern server.
- Password - A password, if required by the specified server.
Running the classification
- On the parameters tab, adjust the parameters, or use the default values as used in this example.
- The first step is generation of the classifier and cross-validation. Click the "Test via Cross-Validation" button.
Progress on the cross-validation is indicated directly in the component.
When the run has completed, the numbers of True and False positives and negatives will be shown under the heading "Cross Validation Results".
- For the second step, the test classification run, now just push the "Analyze" button. When the calculation is complete, the results will be stored in the Arrays/Phenotypes component.
- In this run, 4 of the test arrays were identified as Case, and 8 were identified as control, as shown in the figure below of the Arrays/Phenotypes component.
Example - Weighted Voting (WV)
For the Weighted Voting example we will use the same data setup as described above for KNN. The server settings are also the same.
- Run the classification and cross-validation step as before by pushing "Test via Cross Validation".
- Now hit the "Analyze" button.
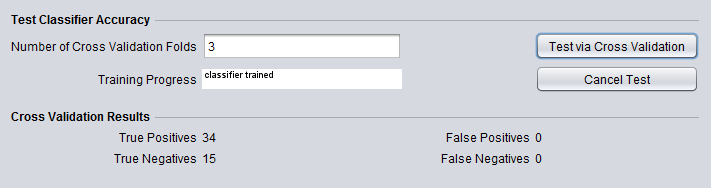
The predicted case and control sets are added to the Arrays component below the results from the KNN analysis.
The result shows that in this case all test arrays were classified as control.
References - GenePattern
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP (2006) GenePattern 2.0 Nature Genetics 38 no. 5 (2006): pp500-501 doi:10.1038/ng0506-500. (PubMed 16642009)
- GenePattern website.
- GenePattern modules documentation.
References - KNN
- Golub T.R., Slonim D.K., et al. (1999) “Molecular Classification of Cancer: Class Discovery and Class Prediction by Gene Expression Monitoring,” Science, 531-537 .
- Slonim, D.K., Tamayo, P., Mesirov, J.P., Golub, T.R., Lander, E.S. (2000) Class prediction and discovery using gene expression data. In Proceedings of the Fourth Annual International Conference on Computational Molecular Biology (RECOMB). ACM Press, New York, pp. 263–272.
- Johns, M. V. (1961) An empirical Bayes approach to non-parametric two-way classification. In Solomon, H., editor, Studies in item analysis and prediction. Palo Alto, CA: Stanford University Press.
- Cover, T. M. and Hart, P. E. (1967) Nearest neighbor pattern classification, IEEE Trans. Info. Theory, IT-13, 21-27, January 1967.
References - WV
- Golub T.R., Slonim D.K., et al. (1999) “Molecular Classification of Cancer: Class Discovery and Class Prediction by Gene Expression Monitoring,” Science, 531- 537.
- Slonim, D.K., Tamayo, P., Mesirov, J.P., Golub, T.R., Lander, E.S. (2000) Class prediction and discovery using gene expression data. In Proceedings of the Fourth Annual International Conference on Computational Molecular Biology (RECOMB). ACM Press, New York, pp. 263–272.