Sequence Retriever
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Overview
The Sequence Retriever component fetches the DNA or protein sequences for selected gene markers from remote databases. Nucleotide sequences are retrieved from the GoldenPath database hosted at the UC Santa Cruz. Amino-acid sequences are retrieved from the European Bioinformatics Institute (EBI).
DNA sequences are retrieved using the transcript start site obtained for the desired gene from the refGenes table at UCSC. This table lists refSeq genes available for each supported species.
The Sequence Retriever component functions only in the context of microarray datasets. The component also depends on the microarray platform annotation file, which must be loaded when the microarray dataset is read into geWorkbench.
For DNA queries, the RefSeq gene(s) associated with a given probeset are obtained from the annotation file and used for the query. For protein queries, UniProt IDs from the annotation file are used. If multiple IDs are present for a marker, they are all used.
One the desired sequences have been downloaded, they can be viewed or saved to the Workspace for use by other components. Examples include running BLAST searches, looking for the presence of known promoter motifs ( Promoter Analysis), or looking for patterns in a set of sequences using Pattern Discovery.
Controls
- Type - DNA or Protein
- Source - The pulldown menu to the right of the "Type" pulldown shows the source to query for the data. At present, only one choice for each type is supported. DNA sequences are retrieved from the UCSC Santa Cruz using RefSeq IDs. Protein sequences are retrieve from the EBI using UniProt IDs.
- Marker - This panel shows markers that are in any activated set in the geWorkbench Markers component.
- Note - If sequences are retrieved for multiple markers, selecting a marker in this list will cause only its hits to be displayed. If no markers are selected in the list, hits for all markers will be displayed (default).
- Find a Marker - This tab allows one to search in the component for a particular marker. However, we recommend performing searches in the Markers component and adding the results to a set.
- "- and +" text fields - For DNA sequence retrieval, these two text fields specify the distance upstream (-) and downstream (+) from the transcript start site for the request. They are disabled when a protein query is selected. The default setting is 2000 bp upstream and 1000 bp downstream.
- Stop - Stop the current sequence download. Only available while a download is in progress.
- Clear - clear the query results display.
- Get Sequence - Launches the query using the selected markers.
- Add to Workspace - Add selected sequences returned by the query to the Workspace.
- View - Line or Full Sequence.
- Show only unique transcript-start sites - Applies to DNA queries only. When checked, only a single sequence entry for a given transcript start site will be shown. If the box is unchecked, all RefSeq sequences associated with the probeset in the annotation file, and for which sequences were retrieved, will be shown.
- Include - Check boxes for sequences to copy to the Workspace when "Add to Workspace" is pushed.
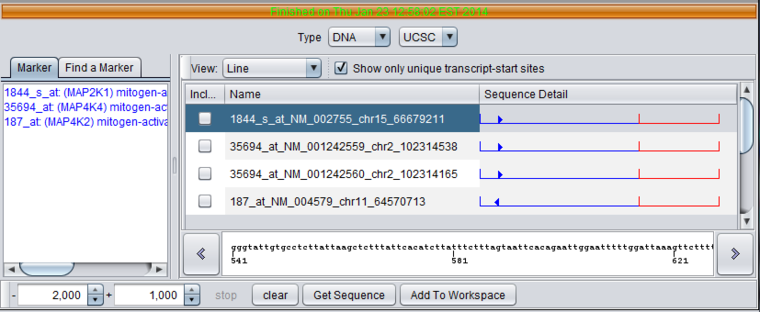
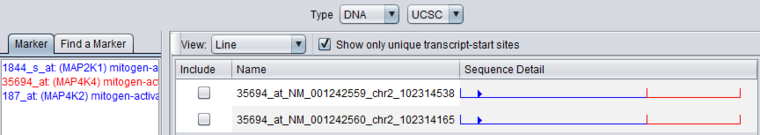
- Name - The name for a sequence is formed by concatenating the marker name with the chromosome number of physical location of the sequence on the chromosome, e.g. in 35694_at_chr2_102314487, "35694_at" is the probeset id, and the sequence is located on chromosome 2 at position 102314487.
- Sequence Detail - graphically depicts the upstream (blue)and downstream (red) sequences areas, and marks the position of the transcript start site and the orientation of the transcript on the genome build.
- right-arrowhead - transcript mapped on genome plus strand.
- left-arrowhead - transcript mapped on genome minus strand.
- Detailed sequence display box - At the bottom of the component, clicking on a sequence in the display above will show the letters with a position scale.
Using the Sequence Retriever
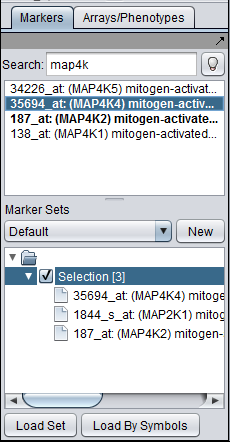
The Sequence Retriever displays a list of the contents of any marker set or sets that are "activated" in the Markers component. "Activated" means that the check-box next to the set's name in the Markers component is checked. These are the markers that will be used in a query to a remote data source.
The Markers component provides a dynamic search feature with which the user can locate desired markers and add them to a set.
DNA or protein sequences can be requested.
For DNA sequences, the Sequence Retriever retrieves genomic sequence from the Golden Path database at the UC Santa Cruz. The amount of sequence upstream and downstream to fetch can be specified in the range settings (-+).
Once sequences have been retrieved, some or all can be added to the Workspace as a new data node ("Add to Workspace" button).
Note - retrieved sequences are not guaranteed to be retained if the marker set using in the query is deactivated, or if the user selects a different data node in the Workspace and then returns to the previous node. One can switch back and forth between DNA and Protein query results within the context of the same marker set.
Prerequisites
- A microarray dataset must be loaded.
- An annotation file must be associated with the microarray dataset at the time it is loaded. At present, only Affymetrix-format annotation files can be read in. These files can be obtained for Affymetrix chip types from affymetrix.com. For exact instructions, please see the geWorkbench FAQ page: FAQ
- A set in the Markers component containing one or more markers of interest must be activated.
- There must be annotation for each marker being queried on. If not, the user will be warned when a marker without annotation is encountered.
Here we show searching for MAP4K4 in the Markers component and adding it to the default "Selection" set by double-clicking. Two other markers have already been added.
When a marker set (here the "Selection" set) is activated, the markers will appear in the Sequence Retriever component.
Retrieving Sequences
- Type - Select DNA or Protein sequences
- Range - If downloading genomic DNA, check/set the range upstream and downstream of the transcript start site for which to retrieve sequence.
- The default range is from -2000 (upstream) +1000 (downstream) relative to the start of the transcript.
- Hit Get Sequence.
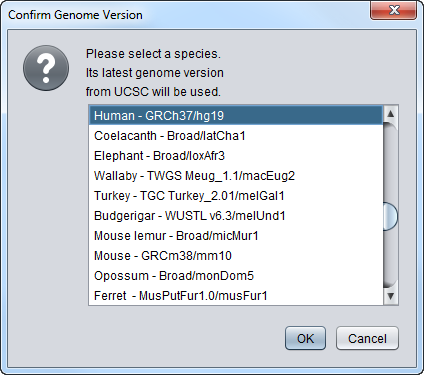
- Select Genome version - If the query is for DNA, the user is prompted to select for the species and version of the genome build to query. You should choose the species corresponding to the microarray chip of the dataset you have loaded. The list of available genome builds is obtained automatically from Santa Cruz.
The retrieved sequences are depicted in the viewer area on the right-hand side of the component. All sequences associated with a given gene symbol are retrieved.
DNA Sequences
DNA Sequence Naming - Each retrieved sequence is given a name formed by concatenating the probeset name for the marker, the refSeq identifier, the chromosome, and the location on the chromosome of the transcript start site obtained from the UCSC refGene table.
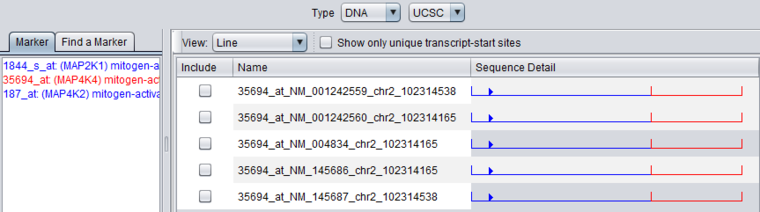
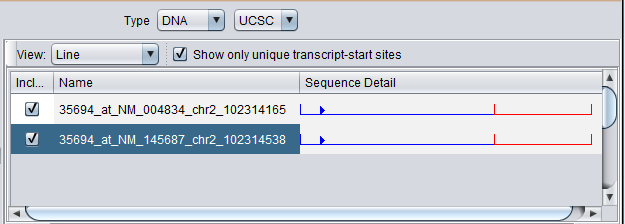
Note - Sequence is only retrieved for the particular RefSeq IDs associated with the probeset in the annotation file. The results may differ from what would be retrieved from UCSC using the gene symbol directly. For example, a direct query to UCSC in this example would return 5 sequences, not just the 2 shown here for this probeset.
Note - If sequences are retrieved for multiple markers, selecting a marker in the list will cause only its hits to be displayed. If no markers are selected in the list, hits for all markers will be displayed.
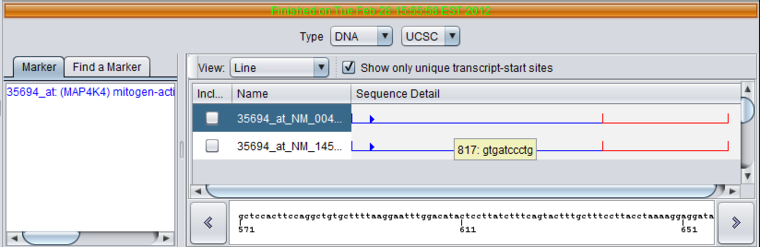
Clicking on a particular sequence under Sequence Detail will display its actual sequence in the scrolling box below.
Selecting just the marker for MAP4K4 in the marker list restricts the display to its hits. For the MAP4K4 marker, genomic sequence for two unique transcript start sites was retrieved.
Unchecking the "Show only unique transcript-start sites" box adds additional sequences with different RefSeq IDs.
Protein Sequences
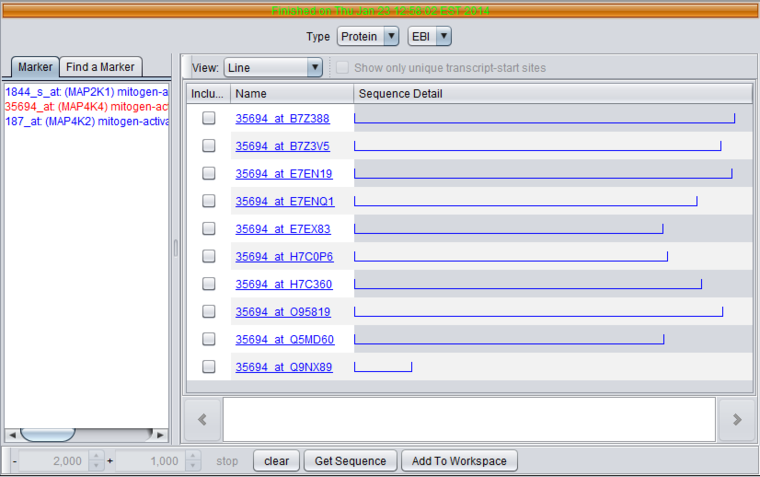
Protein Sequence Naming - For protein results, the sequence name is formed by concatenating the probeset id with the UniProt ID of the sequence.
Note - If sequences are retrieved for multiple markers, selecting a marker in the list will cause only its hits to be displayed. If no markers are selected in the list, hits for all markers will be displayed.
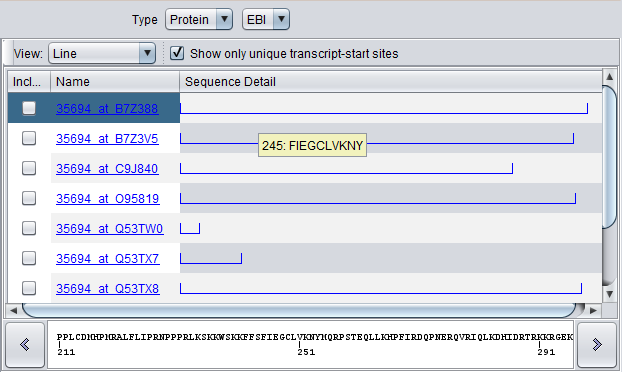
For the MAP4K4 marker, a number of sequences were returned. Note the differences in length as shown graphically.
Viewing Sequences
The graphic sequence viewer pane of the Sequence Retriever component has two viewing modes - "Line" and "Full Sequence", controlled by the "View" pulldown menu.
Line View
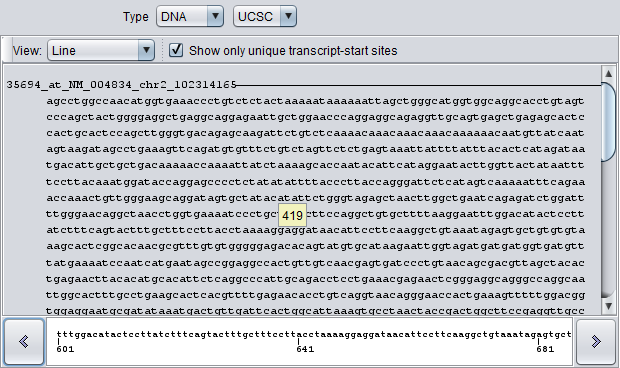
When the view mode is set to "Line", each sequence is represented by a line in the graphic.
Line details for DNA Sequences:
- Blue - for sequence upstream of transcript start site, the line is blue.
- Red - for sequence downstream from transcript start site, the line is red.
- Blue arrowhead - marks the position of the transcript start site and the orientation of the transcript on the genome build.
- right-arrowhead - transcript mapped on genome plus strand.
- left-arrowhead - transcript mapped on genome minus strand.
The line mode has several actions:
- Click - Clicking at any position along a sequence line view causes the local sequence at that position to be displayed in the field at the bottom of the viewer.
- Hover - Placing the mouse cursor over the sequence line view will cause a hover text to appear with the position number and the next 10 sequence letters starting at that position.
- Double-click to Full View - Double-clicking with the mouse on a particular line will cause just that line's full sequence to be displayed in the viewing window.
Double-clicking again in this view will return the display to the line view.
Line view for protein sequences:
For protein sequences, the entire line is blue. Hover and click actions are the same as described for DNA sequences.
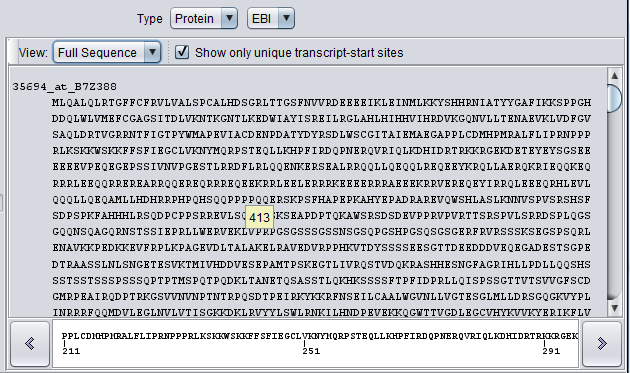
Full Sequence View
When the view mode is set to "Full Sequence", the entire sequence for each marker is displayed.
Saving Sequences to the Workspace
To the left of each retrieved sequence is a check-box, in the column titled "Include". To save sequences to the Workspace, select those desired by checking the adjacent boxes, and then press "Save to Workspace".
All sequences saved at one time will appear in a single new node in the Workspace. You will be prompted to enter a name for the new data sequence node. Here we have given the node the name "MAP4K4_seqs".
Note that when this node is selected, the Viewing area of the geWorkbench GUI will now show components that support working with sequences. However, the Sequence Retrieval component will no longer be visible! You must select the sequence's parent object to see the Sequence Retrieval component again.
Saving sequences to an external FASTA file
Once a group of sequences has been saved to the Workspace, they can also be saved to a file on disk.
- Right-click on the sequence node in the Workspace.
- Select Save.
- Enter a suitable name and save the file.