Component Configuration Manager
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Overview
geWorkbench supports the dynamic loading and unloading of individual components for analysis and visualization. The Component Configuartion Manager (CCM) lists each available component, provides descriptive and license information, and controls loading and unloading. The ability to customize the loaded components may be useful to those who intend only to use particular types of analysis. It may in some cases also increase the responsiveness of the application, as potentially fewer viewer components may need to respond to particular data events (new data set, marker set activation etc.).
In brief, components can be loaded by checking the box adjacent to their entry in the list, and hitting Apply. Components can be unloaded in the same way, by unchecking their box and applying the change.
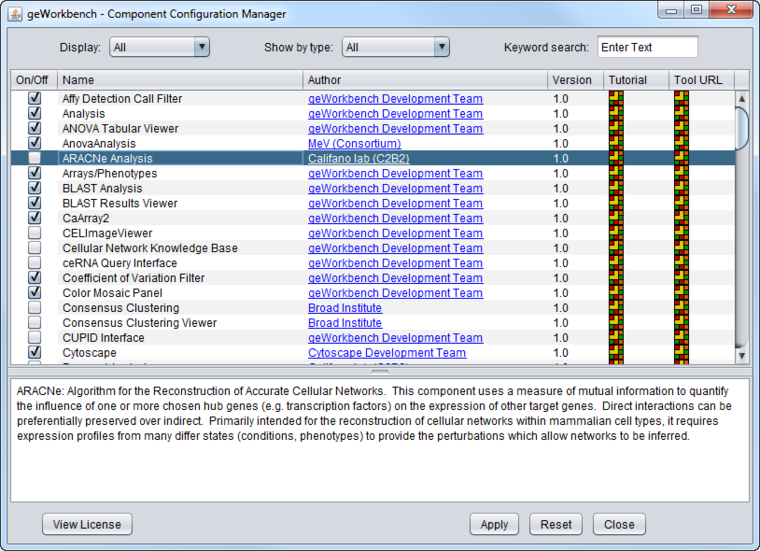
As shipped, the CCM may appear as in the figure below, with only basic components configured. Individual components can be added by clicking on their check boxes.
Note on Workspace saving - The state of the CCM as to which components are loaded is not saved when the Workspace is saved. It is stored separately.
Available Component List
The list of components includes the following column headings:
- On/Off - checking will select the component for loading. Unchecking the box will lead to the component being unloaded.
- Name: Component name
- Author - Component author or source of primary calculation code.
- Version - Component version
- Tutorial - Link to web-based tutorial/manual if available.
- Tool URL - Link to tool home page if available.
Component Dependencies
geWorkbench tracks component dependencies, for example between an analysis component and its viewer. These dependencies are all handled in the CCM.
There are two types of dependency:
- Required - If component A requires component B, then if A is selected for loading in the CCM, B will also be selected. The relationship is reciprocal - if A requires B, then B requires A.
- Recommended - If component B is recommended for use with component A, then if A is selected, the user will be notified that B is recommended. However, component B will not be automatically selected for addition when component A is loaded. The user must select B manually.
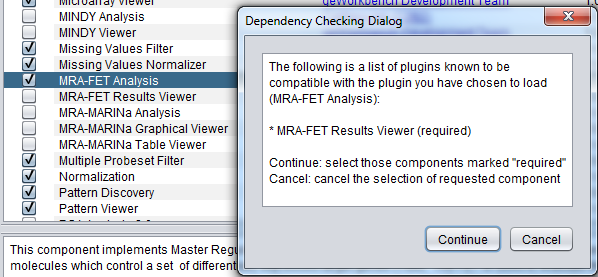
An example of a required relationship is shown in the following figure, where adding the MRA-FET component also requires adding the MRE-FET Results Viewer component.
After clicking "Proceed", both components are selected to be added.
If a component requires agreeing to a license, the license will also be displayed at this time, with the option to accept or reject it.
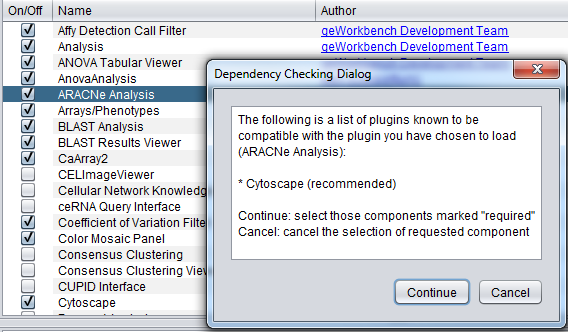
An example of a recommended relationship, between ARACNe and Cytoscape, is illustrated below.
Technical Note
Although the "Continue" button may appear to have focus, hitting Enter may not activate it. However, using the space-bar may work. This is due to cross-platform differences.
Framework Component Dependencies
The Filtering and Normalization framework components have been made dependent on all filter and normalization method components, respectively. Thus these framework components can only be removed if first all filters or normalizers are removed in the CCM.
Controlling display of components
Display
Displayed components can be limited by the following menu choices:
- All
- Only Loaded
- Only Unloaded
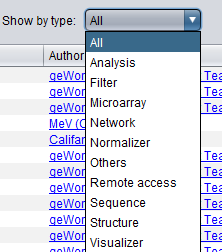
Show by type
In addition, a pulldown menu allows only particular categories of components to be viewed. The categories are not exclusive, a component can belong to multiple relevant categories.
Categories for components include:
- Analysis - data analysis of any kind.
- Filter - microarray data filtering.
- Microarray - microarray data analysis or visualization (does not include filters or normalizers).
- Network - creates, uses or visualizes networks.
- Normalizer - microarray data normalization.
- Others - any components that fall in none of the above categories.
- Remote access - retrieves information from a remote source, e.g. BLAST, sequence retriever etc.
- Sequence - operates on biological sequences.
- Structure - operates on molecular structures (PDB files).
- Visualizer - can visualize data or analysis results.
(Technical Note - The categories are defined dynamically by entries in each component's .cwb file. This file can be edited to add new categories).
Keyword Search
The names of listed components can be searched by typing in a word here. Displayed choices will be progressively narrowed as typing occurs.
Controls
Once the desired changes to the list of components has been made, the changes are put into effect by pushing the Apply button.
- Apply - if any changes have been made to the check-boxes indicating which components are to be loaded, pushing "Apply" causes the changes to actually be be made.
- Reset - If changes have been made but "Apply" has not been pushed, set all checkboxes back to their previous state.
- Close - Close the CCM window.
- View license - If a license is required for a component, it can be displayed by highlighting the component in the list and pressing this button.