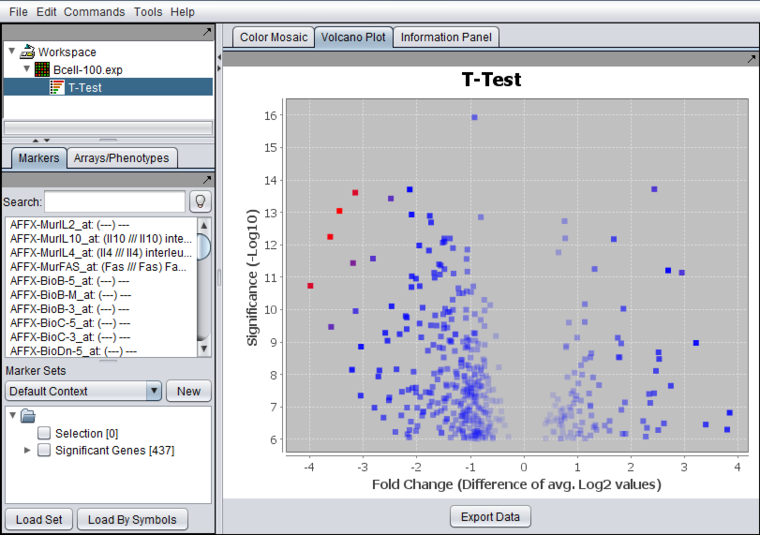
Volcano Plot
Overview
The Volcano Plot graphically depicts the results of the t-test for differential expression. It includes only markers which exceeded the threshold for significance in the t-test. The log2 fold change for each marker is plotted against the -log10 of the P-value.
Markers for which no valid fold-change value could be calculated (e.g. for the case of linear data the average of the case or control values was negative) are omitted from the Volcano Plot. However, all such markers are included if the data is exported to file.
The Volcano Plot viewer is only available when a t-test result is selected in the Workspace.
Fold Change
The method used to calculate fold change depends on whether the data was marked as log2 transformed or not during the t-test using the "Data is log2 transformed" box.
- Linear data ("Data is log2 transformed" box was not checked): the fold change is calculated, for each marker, as the Log2 transform of the average expression in the Case set divided by the average expression in the control set, that is, Log2(Avg(cases)/Avg(controls)) or Log2(Avg(cases)) - Log2(Avg(controls)). (Difference of logs of averaged values).
- Log2 transformed data ("Data is log2 transformed" box was checked): In this case, for each marker, the average of the (log) case values minus the average of the (log) control values is calculated, that is, Avg(cases) - Avg(controls). (Difference of averaged log values).
Color scheme
The lower 2/3 of the absolute values of the (fold change) * (significance) are colored in shades from light blue (lowest values) to dark blue (highest values). The highest 1/3 of such values are colored from dark blue (lowest values) to red (highest values).
Export Data
Exports data for all markers in the t-test result node to a CSV format file.
The column headers are: Probe Set Name, Gene Name, p-Value, Fold Change (log2).
Markers found significant in the t-test, but for which a fold change value could not be calculated, are included in the export file as "N/A". They are not displayed in the Volcano Plot.