Hierarchical Clustering
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Overview
For the geWorkbench web version of Hierarchical Clustering please see Hierarchical_Clustering_web.
Hierarchical clustering is a method to group arrays and/or markers together based on similarity of their expression profiles.
geWorkbench implements its own code for agglomerative hierarchical clustering. Starting from individual points (the leaves of the tree), nearest neighbors are found for individual points, and then for groups of points, at each step building up a branched structure that converges toward a root that contains all points. The resulting graph tends to group similar items together.
Results of hierarchical clustering are displayed in the Dendrogram component, which is further described below.
Prerequisites
Dataset
A microarray dataset must be loaded in the Workspace. If an annotation file is also loaded corresponding to the microarray type, then gene names will be used in the results display, otherwise probeset names will be used.
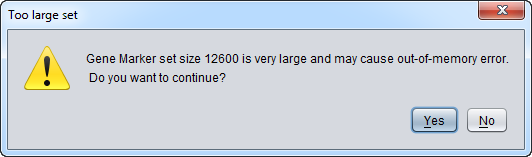
Note - hierarchical clustering is memory intensive. With the default memory settings (see here to change), clustering more than about 2000 markers is not recommended.
If more than 1000 markers or 1000 arrays are selected for clustering, a popup warning will be issued.
The actual number of markers or arrays that can be clustered depends on the amount of memory allocated to the Java virtual machine on your computer.
Missing Values
If there are any missing values in the dataset, an error message will be returned if Hierarchical Clustering is run. Missing values can be filtered out or replaced using the Missing Value Filter or the Missing Value Normalizer.
Parameters
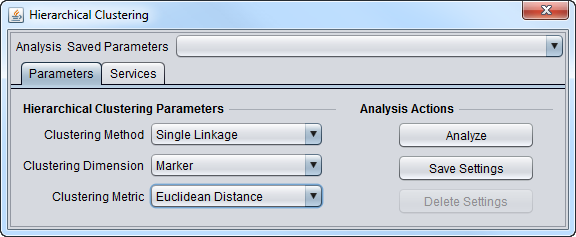
Clustering Method
This parameter is used to indicate the convention used for determining cluster-to-cluster distances when constructing the hierarchical tree. Available options are:
- Single Linkage - The distances are measured between each member of one cluster each member of the other cluster. The minimum of these distances is considered the cluster-to-cluster distance. This method often leads to a "chaining" effect and is usually not recommended.
- Average Linkage - The average distance of each member of one cluster to each member of the other cluster is used as a measure of cluster-to-cluster distance.
- Total Linkage - The distances are measured between each member of one cluster each member of the other cluster. The maximum of these distances is considered the cluster-to-cluster distance.
Clustering Dimension
These are used to indicate whether to cluster markers, microarrays, or both.
- Marker - Cluster the selected markers (genes) based on the similarity across microarrays.
- Microarray - Cluster the selected microarrays based on the similarity across markers.
- Both - Cluster both markers and microarrays.
Clustering Metric
The values being clustered, whether markers or microarrays, can each be represented by vectors of numbers, essentially either rows (markers) or columns (microarrays) taken from a spreadsheet view of all expression values. Several methods by which to calculate the distance between any two vectors are offered:
- Euclidean - The direct, point-to-point distance is calculated (square root of the sum of square differences).
- Pearson's - Pearson's correlation coefficient for two vectors is calculated.
- Spearman's - Spearman's rank correlation coefficient for two vectors is calculated.
Set Selection
Activating one or more sets of markers or arrays in the Marker_Sets or Array_Sets components limits an analysis to those items in the active set(s).
Analysis Actions
This component uses the standard analysis component framework, which provides three buttons:
- Analyze - Start the clustering job.
- Save Settings - Save the current settings to a named entry in the settings list.
- Delete Settings - Delete the selected setting entry from the list.
Services (Grid)
Hierarchical Clustering can be run either locally within geWorkbench, or remotely as a grid job on caGrid. See the Grid Services section for further details on setting up a grid job.
Running Hierarchical Clustering
This example clusters a set of markers generated through a t-test. You can generate or select any set of markers to run your own example.
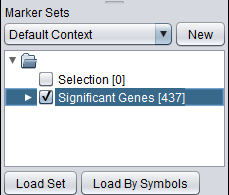
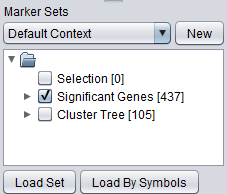
1. Activate a set of markers. Here, "Significant Genes [437]" ( which contains 437 markers) was activated.
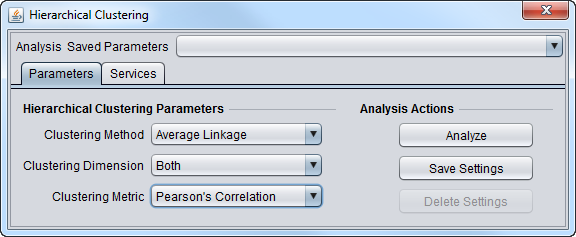
2. Set the parameters as desired. Here, we used:
- Clustering methods: Average Linkage.
- Clustering Dimension: Both.
- Clustering Metric: Pearson's Correlation.
3. Click Analyze.
4. A progress bar will be visible during the calculation, first displaying a message regarding computing distances....
... and then about clustering.
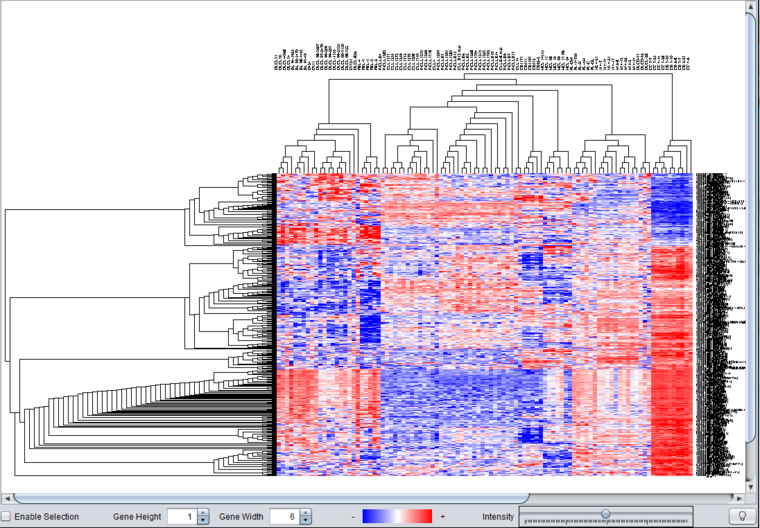
The results are placed in the Workspace and labeled "Hierarchical Clustering", and are displayed in the Dendrogram component.
The Dendrogram Visual Component
Hierarchical clustering results are displayed in the Dendrogram component.
Controls
Enable Selection
Checking this box allow a subtree of the dendrogram to be selecting interactively using the mouse. The selected area is highlighted in blue. Clicking on the selected area will restrict the display to just the selected portion of the tree.
Gene Height
Sets the height in pixels of the rows devoted to each marker and associated labels.
Gene Width
Sets the width in pixels of the columns devoted to each array. Label text is also scaled proportionately.
Color key
Shows the range of color values from lowest to highest expression for the current display preference. See the Color Mosaic tutorial for further details on the absolute and relative display preference settings.
Intensity slider
The intensity slider adjust the midpoint of the color scale to lower or higher expression values.
Bulb Icon
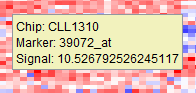
Pushing the bulb icon activates a tool-tip feature on the dendrogram display. Mousing over the dendrogram will bring up a display of the following information for any point:
- Chip: the array name
- Marker: the marker (probeset) name.
- Signal: the expression value of the selected marker on the selected array.
Right-clicking on the dendrogram will show a menu with two entries:
- Image Snapshot - place a static snapshot of the tree as currently displayed into Workspace.
- Add to Set - add the markers represented in the currently displayed tree to a new set in the Markers component called "Cluster Tree". This is most useful if done after a subtree of markers has been selected.
Working with Hierarchical Clustering Results
Selecting a subtree
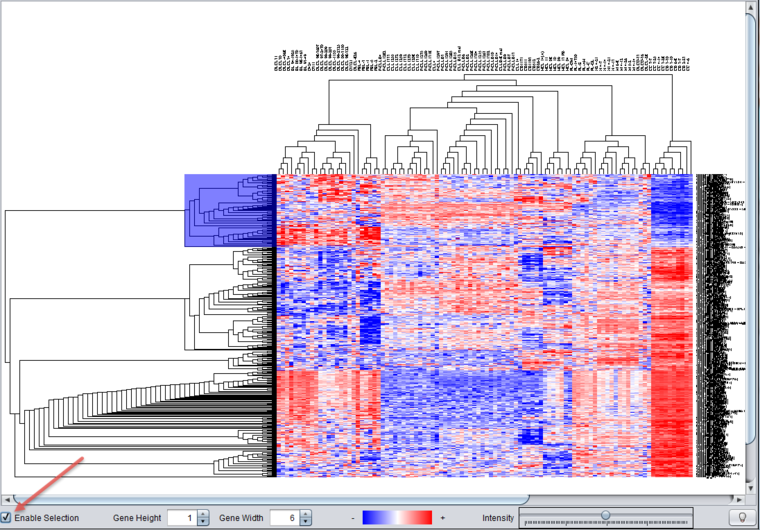
The Dendrogram component allows one to select and work with just a portion of the displayed tree. To activate this feature, check the Enable Selection checkbox at lower left in the Dendrogram component (indicated by the red arrow). The subtree selection will work for both markers and arrays, depending only on if they were included in the initial clustering calculation.
The following figure illustrates selecting a subtree of markers. Moving the cursor over the displayed tree draws a blue rectangle over the selected portion.
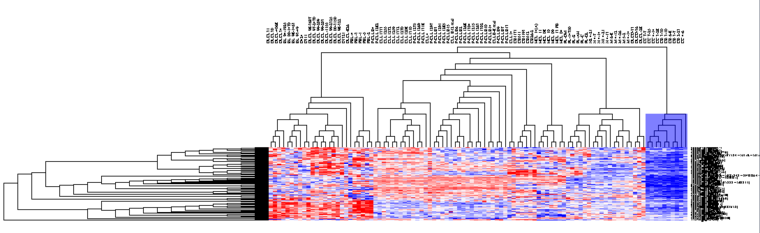
Clicking on the selected area will cause only this area to be displayed, as shown below. A subset of arrays can also be selected, again shown in blue highlighting.
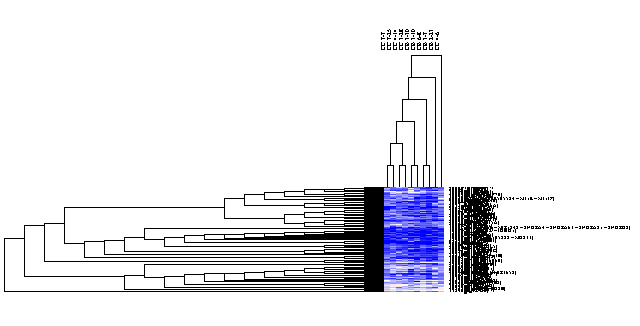
The result of the second selection is this small set of data points:
Working with a subtree in the Dendrogram
As already mentioned, the right-click menu allows one to save the markers in a displayed subtree to the Markers component, or the arrays in a displayed array subtree to the Arrays/Phenotypes component:
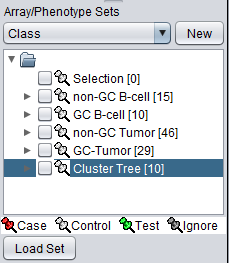
- Add to Set - add the markers and arrays represented in the currently displayed tree to new sets in the Markers and Arrays components called "Cluster Tree".
The following figure shows the the new set of markers (labeled "Cluster Tree [105]") after it has been added to the Markers component. The number in brackets indications how many markers are contained in the set.
Arrays are added to the Arrays component (labeled "Cluster Tree [10]"), as shown here: