File Formats
Contents
Affymetrix MAS5/GCOS Format
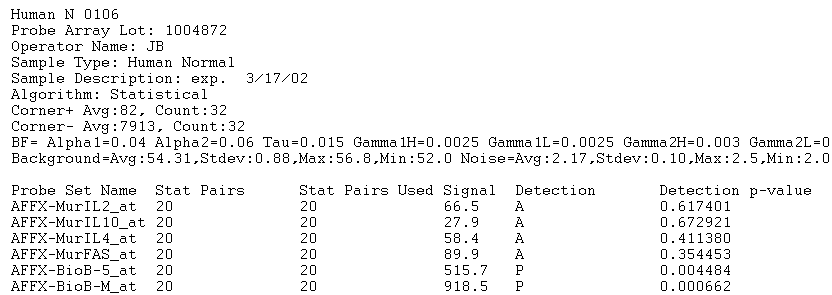
These are text files (specifically, text versions of .CHP files) produced by the MAS software from Affymetrix. The image below provides an example of an input file that the application will accept as a correctly formatted "Affymetrix MAS5/GCOS" file type (only the first 18 lines of the file are shown):
Any number of lines can precede the actual array data (in the example above the first 11 lines are non-data lines). All such lines will be ignored when the file is parsed. The beginning of the actual data is marked by a row containing tab-separated column names. Any number of columns may be present; however, only the following columns will be acted upon (column names must be spelled exactly as listed below and cannot contain tabs):
- Probe Set Name: This column is mandatory; the file will fail to be parsed unless this column is present. Its contents are strings that provide the marker names associated with a microarray set (AFFX-BioB-5_at, AFFX-MurIL10_at, etc).
- Signal Log Ratio: This column is optional. If present, its contents must be real numbers and will be used as the expression measurements for the corresponding markers.
- Signal: Same as column "Signal Log Ratio" above. If both columns are present, only "Signal Log Ratio" is used ("Signal" will be ignored)
- Avg Diff: Same as columns "Signal Log Ratio" and "Signal". If either "Signal Log Ratio" or "Signal" is present then "Avg Diff" is ignored and the contents of those columns are used instead (with "Signal Log Ratio" taking priority over "Signal"). At least one of these 3 columns ("Signal Log Ratio", "Signal", "Avg Diff") must be present for the file to parsed in a meaningful way. if this is not the case, the file will still be read in but all markers will be tagged as having missing values.
- Detection: This column is optional. Its value is a single character (P, M, or A) and if present it is used to determine the absolute call for a measurement (A -> absent, M -> marginal, P -> present).
- Abs Call: Exactly the same as column "Detection" (for compatibility with text files produced by older versions of the MAS software).
- Detection p-value: This column is optional. Its values are real numbers between 0 and 1. It provides a measure of confidence in the quality of the measurement reading (the smallest the p-value the higher the confidence). Further, if both "Detection" and "Abs Call" columns are missing, the "Detection p-value" is used as an alternative method to determine the absolute calls for marker measurements:
- If p-value < 0.04, then the absolute call is "present"
- If 0.04 <= p-value < 0.06, then the absolute call is "marginal".
- If p-value >= 0.06, then the absolute call is "missing".
The p-value thresholds used above are chosen to be the same as the default thresholds used for the same purpose by the MAS software (see the manual "GeneChip Expression Analysis: Data Analysis Fundamentals" from Affymetrix, http://www.affymetrix.com/support/downloads/manuals/data_analysis_fundamentals_manual.pdf).
The marker data follow the column names. Each row correspond to one marker. Values within a row are tab-separated.
Affymetrix File Matrix Format (geWorkbench ".exp" format)
The "EXP" format file is the geWorkbench native format for saving microarray data. It allows not only the data matrix for a group of arrays to be saved, but also various set labels grouping these arrays, e.g. by phenotype. The data is saved in a tab-delimited spreadsheet format.
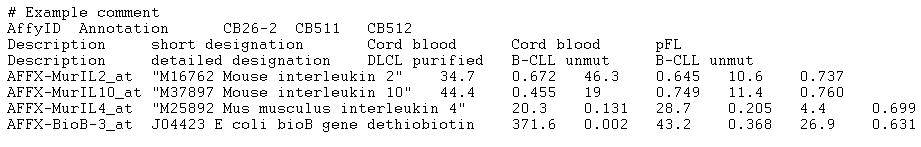
Below are the first few lines of a "Affymetrix File Matrix " file type:
This example shows data from 3 microarrays named CB26-2, CB511 and CB512).
Any number of comment lines, each one starting with a #, can precede the actual array data (however, no empty lines are allowed before, after or between comment lines). All comment lines will be ignored during file parsing.
The beginning of the actual data is marked by the first row that does not begin with a # character. This row is expected to have N+2 tab-separated column names, where N is the number of microarrays. The first column name must read "AffyId"; the second column name must read "Annotation". The subsequent N entries are assumed to be the labels of the N microarrays whose data are contained in the file. Any string can be used as a microarray label as long as it does not contain tab characters.
There can then follow zero or more "Description" lines. Description lines can be used to group microarrays into array sets - see the tutorial Data Subsets - Arrays". Each line describes one list of array sets and comprises N+2 tab separated entries. The first entry must be the word "Description". The second entry is the label that will be used in the application to identify this list of array sets (users can select which list to work with from the "Array/Phenotype Sets" drop-down menu within the Arrays/Phenotypes application component). The following N columns are array set labels that map one-to-one to the N arrays listed in the "AffyID" line; their relative order in the description line determines how the arrays are grouped into sets. E.g., in the example above the first description line defines a list containing 2 array sets: the first set is called "Cord blood" and contains the arrays CB26-2 and CB511. The second set is called "pFL" and contains the array CB512. The list with these two array sets is itself named "short designation".
Within a particular array list, an array can belong to more than one set. In the cell of the tab-delimited file for such an array, each set that the array is a member of is separated by the "|" pipe character. For example, "a|b|c" would assign the array to sets a, b and c.
The last part of the file contains the actual expression measurements. There is one line per marker on the chip (there are 4 such lines in the example). Each line starts with the name of the marker. The name is followed by a string that provides a human readable annotation of the marker. The measurement values follow the annotation string. This portion of a data line can assume one of two possible forms:
- Measurements and p-values: there are 2N tab separated decimal values present (the example above has this form). These values are parsed in consecutive pairs. The i-th pair provides data for the value of the marker on the i-th array and comprises (a) the expression measurement for the marker on the array and (2) a p-value (between 0 and 1) indicating the strength of the call. The p-value is used by the application to infer the detection call for the corresponding measurement. The algorithm used for this purpose is the same as the one used in the Affymetrix MAS5/CCOS Format to infer detection calls from the value of the "Detection p-value" column.
- Measurements only: there are N tab separated decimal values present. The i-th values provides the expression measurement for the marker at the i-th microarray. All measurements are treated as having a detection call of "Present".
- Missing Values - As of release 2.5.0, missing values (no entry at all between two tabs) are allowed in the data lines of the file. Previously, some entry was required, if just a space. The number of arrays and the number of data cells must agree (x2 if p-values are used).
Notes
- In the case where there are two data values per array, e.g. signal and p-value, the format is difficult to create by hand because the header columns do not directly label or line up with the data columns.
- Example file: An example of the first 14 lines of the file Bcell-100.exp (used in many examples in these tutorials) is available here.
Tab-delimited format
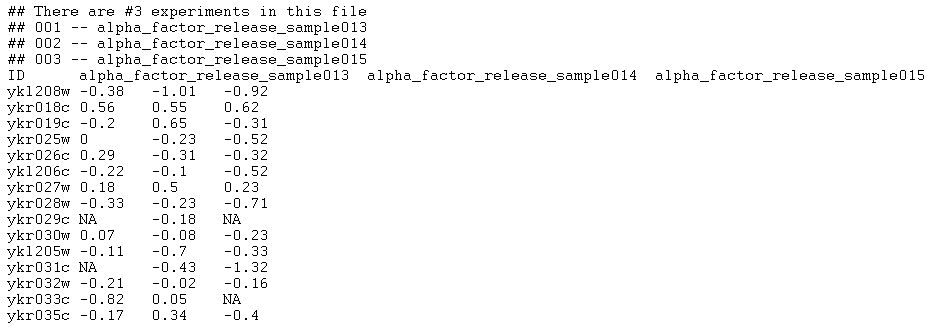
This is an example of a file that the application will accept as a correctly formatted "Tab-delimited " file type (only the first 20 lines of the file are shown), as output by the program RMAExpress:
Any number of comment lines, each one starting with a # or a !, can precede the actual array data (however, no empty lines are allowed before, after or between comment lines). All comment lines will be ignored during file parsing. The beginning of the actual data is marked by the first row that does not begin with a # or a ! character. This row is expected to have N+1 tab-separated column names. The first column name can be any character (in the example above it has the value "ID"). The first column name cannot be left blank or be occupied with a white space (for example, a header cannot start with "\t"). The remaining N entries are assumed to be the names of the microarrays comprising the microarray set (in the example above the microarray names are "alpha_factor_release sample013", "alpha_factor_release_sample014" and "alpha_factor_release_sample015").
Subsequent lines contain the actual data. Each line corresponds to a single marker and consits of N+1 tab-separated entries. The first entry is the marker name (a string). The remaining N entries are real numbers providing the expression level of the marker at each of the microarrays in the set. A missing value or a non-real value results at a measurement being marked as "missing".
Notes
- This simple tab-delimited format just contains array and marker names and the expression data. It does not contain any annotation, nor does it support any groupings of arrays.
- This format only supports a single value per marker per array. A second, confidence value is not supported.
- Some microarray platforms can include multiple markers/probesets for some or many of the genes represented. If a data file contains e.g. gene symbols rather than individual marker names in the first column, any resulting duplicate appearance of such a label will prevent the file from being read in to geWorkbench.
GenePix Format
Each GenePix file contains data for a differential expression experiment involving a single microarray. A detailed description of the GenePix .gpr format can be found at the manufacturer's customer service site.
The top portion of the file (the GPR Header) contains information about image acquisition and analysis. This part is ignored during parsing. From the actaul data portion of the file only the following data columkns are parsed:
- ID: The contents of this column are used as the marker names.
- F635 Median, F635 Mean, B635 Median, B635 Mean, F532 Median, F532 Mean, B532 Median, B532 Mean: The values of these columns are used for calculating the composite expression measurement for a marker. There are 4 possible options for performing this calculation (the Tools->Preferences menu can be used to specify which of the 4 to use):
- ( F635 Mean - B635 Mean) / (F532 Mean - B532 Mean).
- ( F635 Median - B635 Median) / (F532 Median - B532 Median).
- ( F532 Mean - B532 Mean) / (F635 Mean - B635 Mean).
- ( F532 Median - B532 Median) / (F635 Median - B635 Median).
- Flags: Parsed as an arbitrary string. The Flags value associated with a marker can be used to filter out the marker when using the "Genepix Flags" filter (for details see the online help section on Filters).
MAGE-TAB Data Matrix Files
Please see the entry MAGE-TAB Data Matrix Files in the Local Data Files chapter.
An example of the supported data matrix format is found here.
Annotation Files
As of geWorkbench 2.4.0, geWorkbench supports two Affymetrix annotation file types, in CSV (comma separated value) format:
- Affymetrix 3' Expression
- Affymetrix Gene/Exon 1.0 ST
Prior to geWorkbench 2.4.0, geWorkbench supported only the Affymetrix 3' Expression annotation file format.
In these annotation files, rows represent individual probesets, and each column contains a different annotation type.
Affymetrix annotation files for their platforms can be obtained directly from the Affymetrix support website (registration required). These files are updated quarterly.
geWorkbench expression file parser support for Affymetrix annotation files
These geWorkbench file parsers will accept an Affymetrix-format annotation file during the data loading process:
- Affymetrix file matrix (geWorkbench native format)
- MAS5/GCOS (Affymetrix)
- GEO SOFT
- GSM - sample
- GSE - series, a series of samples representing an experiment
- GDS - data set, a curated data matrix
- GEO Series Matrix
- MAGE-TAB data matrix
- Tab-delimited (e.g. RMAExpress, etc).
The geWorkbench GenePix file parser does not support a separate annotation file.
Creating a custom annotation file
For a non-Affymetrix platform, you can create your own custom annotation file using only the annotation columns needed for the analyses you intend to perform in geWorkbench. It is easiest to use the Affymetrix 3' Expression file format. Note that for working with Gene Ontology analysis, GO term annotation for each marker must be provided.
The geWorkbench annotation file parser recognizes the column headers shown in the next section. All columns are optional, however, if "probeset id" is not present, none of the annotation records will link to the expression file.
Recognized Column Headers for Affymetrix 3' Expression
- "Probe Set ID"
- "Species Scientific Name"
- "Archival UniGene Cluster"
- "UniGene ID"
- "Genome Version"
- "Alignments"
- "Gene Title" (e.g. Epidermal growth factor receptor)
- "Gene Symbol" (e.g. EGFR)
- "Entrez Gene"
- "SwissProt"
- "RefSeq Transcript ID"
- "Gene Ontology Biological Process"
- "Gene Ontology Cellular Component"
- "Gene Ontology Molecular Function"
- "Pathway"
- "Transcript Assignments"
For fields that contain multiple values, the delimiter between the values is "///".
Parsing Errors
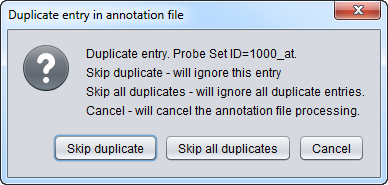
The geWorkbench parser will check for whether a given probeset occurs more than one time in the annotation file. If it does, an error dialog will appear, offering three choices:
- Skip this duplicate annotation entry.
- Skip all duplicate annotation entries.
- Cancel - do not load the annotation file.
This dialog is shown in the following image:
Technical Information
Addition of support for the Affymetrix Gene/Exon 1.0 ST file type was described in Mantis issue #3006 and #3027. In particular, to create a custom version using that format, the 5 sub-fields of the gene_assignment column must be populated. The Affymetrix parser was modified (Mantis issue #3053) to properly handle an annotation file that has a superset of the markers actually present in the microrray dataset.
Network Formats
Tools such as ARACNe, the CNKB, and Master Regulator Analysis (MRA) make use of network interaction files. ARACNe creates "adjacency matrix" (ADJ) format files, and MRA reads them. The CNKB can export complete sets of interactions in either the ADJ or SIF format.
SIF format
The Simple Interaction Format (SIF) was developed for Cytoscape.
For a full definition see the Cytoscape manual, for example Ctyoscape manual v2.8
Each line contains interactions of a particular type for the first node with one or more target nodes:
- node1 interaction-type-code node2 node3 node4 etc.
Some interaction-type-codes used in the CNKB are
- pp protein-protein
- pd protein-DNA
- tm modulator-TF
Adjacency matrix (ADJ) format
The is is format used by ARACNe.
The first entry on a line is a marker used as a "hub" in the ARACNe calculation, e.g. node1 in the example below.
For each of the interactions in which hub1 takes part, target markers (e.g. target1 , target2) and a weight for each such interaction (e.g. value1, value2) are listed.
hub1 target1 value1 target2 value2 target3 value3 etc.... hub2 target4 value4 target5 value5 target6 value6 etc....
The weight, valueN can be for example the mutual information, a confidence value etc. When generated by ARACNe, the value is the mutual information between the hub and the target. The adjacency matrix generated by ARACNe only contains hub-target interactions that exceed a threshold level.
"Lab Format"
Some tools can import networks in "Lab Format". This format is a five column, tab-delimited file with the following column headers and content types:
- Gene1 - identifier for gene 1
- Gene2 - identifier for gene 2
- pdi - boolean: 1 if interaction is protein-DNA, 0 otherwise
- ppi - boolean: 1 if interaction is protein-protein, 0 otherwise
- Dir - boolean: 1 if interaction is directed, 0 otherwise
Example, in terms of EntrezIDs:
Gene1 Gene2 pdi ppi Dir 1019 3320 0 1 0 1025 3320 0 1 0 1147 3320 0 1 0 1385 3320 1 0 1 1445 3320 0 1 0
Network alternative "5-column file format"
Each line represents a network edge and comprises five tab-delimited columns:
- Transcription factor id: A string that provides the transcription factor end of the edge. This is usually a probeset id or a gene symbol.
- Target id: A string that provides the target end of the edge. This is usually a probeset id or a gene symbol
- Mutual information: The mutual information (MI) of the edge (a real number). If the edge MI is not known/available, the value 1 can be entered here.
- Spearman's correlation: The Spearman's correlation for the edge, computed on the original microarray set that gave rise to the ARACNe network (a real number). If not known/available, the value 1 can be entered here.
- P-value for Spearman's correlation. The p-value associated with Spearman's correlation found in the previous column (a real number between 0 and 1). It this p-value is not known/available, a value of 0 can be entered here.
Misc. Formats
MRA Export format
The data is saved in blocks of comma separate values, one block per "master regulator". The first line of each block is the marker id and the gene name of the master regulator. Subsequent lines contain, in addition, a zero, and then the t-value for that markers. The position with zero previously contained a p-value which is no longer calculated.
220462_at, CSRNP3 200660_at, S100A11, 0.0, 8.681083 201474_s_at, ITGA3, 0.0, 6.0778093 202910_s_at, CD97, 0.0, 7.482091 203370_s_at, PDLIM7, 0.0, 7.235049 205479_s_at, PLAU, 0.0, 9.105933 208540_x_at, S100A11, 0.0, 8.539139 208690_s_at, PDLIM1, 0.0, 7.2551293 210735_s_at, CA12, 0.0, 6.954225 211924_s_at, PLAUR, 0.0, 6.1465526 214853_s_at, SHC1, 0.0, 9.554302
202614_at, SLC30A9 160020_at, MMP14, 0.0, 4.3422446 200808_s_at, ZYX, 0.0, 6.7016177 200859_x_at, FLNA, 0.0, 6.5627837 201315_x_at, IFITM2, 0.0, 6.2328486 201389_at, ITGA5, 0.0, 6.451033 201883_s_at, B4GALT1, 0.0, 3.3089304 202888_s_at, ANPEP, 0.0, 4.060898 203149_at, PVRL2, 0.0, 4.0649285 203370_s_at, PDLIM7, 0.0, 7.235049 205936_s_at, HK3, 0.0, 3.5691133 207667_s_at, MAP2K3, 0.0, 5.186854 208690_s_at, PDLIM1, 0.0, 7.2551293 209359_x_at, RUNX1, 0.0, 6.7081113 211924_s_at, PLAUR, 0.0, 6.1465526 211966_at, COL4A2, 0.0, 7.394549 214752_x_at, FLNA, 0.0, 7.3759694 214866_at, PLAUR, 0.0, 6.628238 215498_s_at, MAP2K3, 0.0, 7.774499 215706_x_at, ZYX, 0.0, 6.2943435 217691_x_at, SLC16A3, 0.0, 5.4462223 221807_s_at, TRABD, 0.0, 3.2993512 35626_at, SGSH, 0.0, 6.4077697
Pattern Discovery Export Format
- pattern number
- pattern
- [parameters]
- [sequence number, starting hit position in sequence] (both numbers are zero-based)
discovery File:C:\Users\ksmith\Desktop\geWB_data\fasta\H1H5_HistoneDB_NHGRI.fasta [0] LKER.GSS [4,4,7,209.92124911313488] [0,57][1,67][2,68][4,59] [1] L.QTKG.GASGSFKLS [3,3,14,604829.8848207161] [0,100][3,94][4,102] [2] A.AKKP.AK [4,4,7,172.99810658850754] [0,239][1,225][2,225][4,245] [3] AT.KKP.AK [4,4,7,172.99810658850754] [0,239][1,132][2,133][4,245] [4] KKP.AKKA [3,3,7,11.941700055286251] [1,228][2,228][4,165] [5] AA.KKA.AAA [3,3,8,54.52686758911383] [1,193][2,194][3,61]
Array Set File Format
Array sets can be loaded from disk into the Arrays component.
The file can contain one or two columns.
- The first column contains array names corresponding to those in the current expression data set.
- The optional second column can contain set names. Each array will be assigned to the indicated set on read-in. If a set does not already exist, it will be created.
Marker Set File Format
Marker sets can be loaded from disk into the Markers component.
- The file has a single column containing either probeset names or gene names.