Consensus Clustering
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Overview
This component allows geWorkbench to run Consensus Clustering on a GenePattern server. The user must supply the URL of an available GenePattern server. All results are returned to geWorkbench, and the clustered arrays or markers are available as sets for further analysis.
As described in the GenePattern documentation, "Consensus clustering provides for a method to represent the consensus across multiple runs of a clustering algorithm and to assess the stability of the discovered clusters. To this end, perturbations of the original data are simulated by resampling techniques".
For further information on the Consensus Clustering algorithm, please see the GenePattern documentation at GenePattern Analysis:Modules.
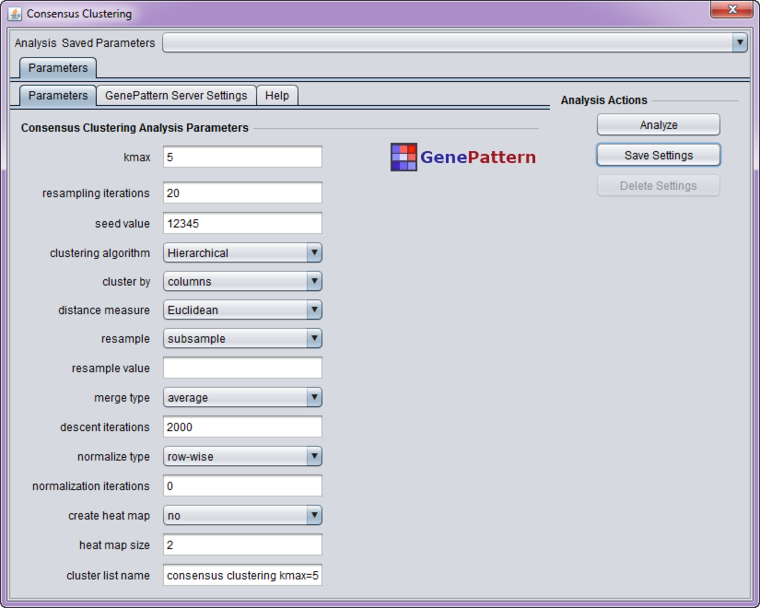
Parameters
- kmax - Try K=2,3,...,kmax clusters (must be > 1)
- resampling iterations - Number of resampling iterations
- seed value - Random number generator seed
- clustering algorithm - Type of clustering algorithm. Choices are
- Hierarchical
- SOM
- NMF
- KMeans
- cluster by - Whether to cluster by rows/genes or columns/experiments
- distance measure - Distance measure. Either
- Euclidean
- Pearsons
- resample - resampling scheme (one of 'subsample[ratio]', 'features[nfeat]', 'nosampling')
- resample value - ratio or nfeat can be optionally input as indicated above.
- merge type - Ignored when algorithm other than hierarchical selected
- Average
- Complete
- Single
- descent iterations - Number of SOM/NMF iterations
- output stub - Stub prepended to all the output file names
- normalize type - row-wise, column-wise, both, none.
- normalization iterations - number of row/column normalization iterations (supersedes 'normalize type')
- create heat map - Whether to create heatmaps (one for each cluster number): no or yes
- heat map size - point size of a consensus matrix's heat map (between 1 and 20)
- cluster list name - name for new geWorkbench marker/array context in which to store clusters. This should be changed to a name meaningful for your setup.
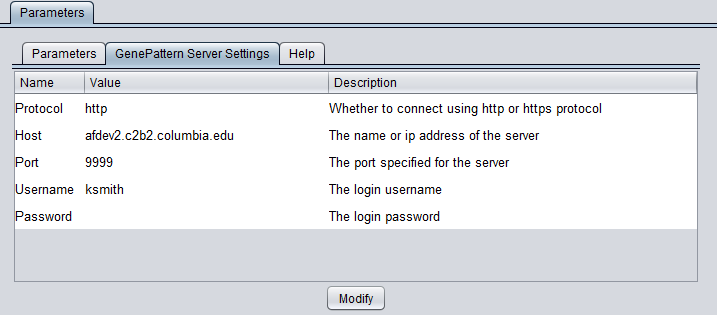
GenePattern Server Settings
You can connect to any running GenePattern server to run the analysis (provided it has the required module installed). An example configuration of the "GenePattern Server Settings" tab is shown here:
To run GenePattern components, a GenePattern account is required.
Pushing "Modify" brings up an editing box where any of the settings can be changed.
- Protocol - HTTP or HTTPS, depending on the server being used.
- Host - URL of a GenePattern server.
- Port - Port at which the GenePattern server is located on the Host machine.
- Username - A valid user name on the specified GenePattern server.
- Password - A password, if required by the specified server.
Results
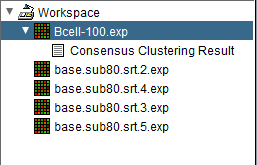
After the Consensus Clustering algorithm completes, the result for each value of kmax is placed into the Workspace as a new data matrix file. The picture below shows the four new data matrix nodes in addition to the original dataset (highlighted).
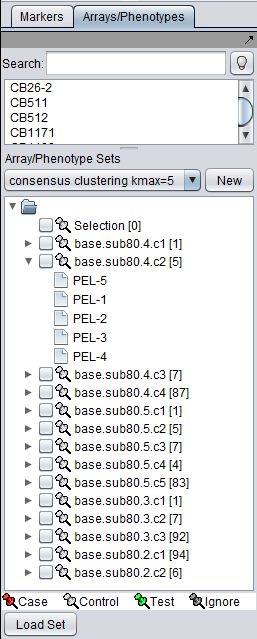
The sets of arrays or markers belonging to each cluster in a particular result are stored in a new Array or Marker context of the parent data node. The picture below shows the array sets placed into the Markers/Arrays component. One of the sets has been opened to show the 5 array members.
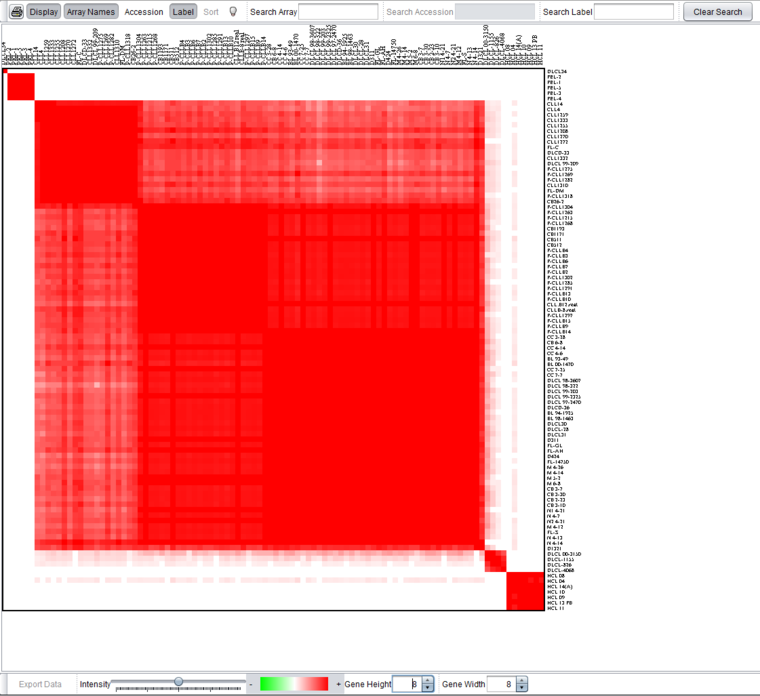
In the figure below, the data node resulting from a run with kmax=5 (base.sub80.srt.5.exp) has been selected in the Workspace. The clustering result can be viewed in the Color Mosaic component as shown below.
Access to Original Output Files
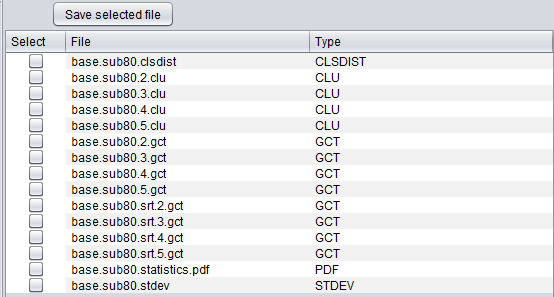
A special file browser is also provided, which can be activated by selecting the "Cluster Results" node under the parent data node in the Workspace. Any desired result file can be selected and exported to disk in its native format. The files returned include the clusters represented in the GenePattern GCT format, the cluster statistics files, and individual clusters.
- Select - check the box for files to be saved to disk.
- Save selected file - save the selected files to disk.
References - GenePattern
- Reich M, Liefeld T, Gould J, Lerner J, Tamayo P, Mesirov JP (2006) GenePattern 2.0 Nature Genetics 38 no. 5 (2006): pp500-501 doi:10.1038/ng0506-500. (PubMed 16642009)
- GenePattern website.
- GenePattern modules documentation.
References - Consensus Clustering
Monti S, Tamayo P, Mesirov J, and Golub T. Consensus Clustering: A resampling-based method for class discovery and visualization of gene expression microarray data (2003) Machine Learning Journal 52(1-2):91-118.