Difference between revisions of "GSEA"
(→Prerequisites) |
(→References) |
||
| Line 52: | Line 52: | ||
* [http://www.broadinstitute.org/cancer/software/genepattern/modules/docs/GSEA/14 GSEA v14] online documentation. | * [http://www.broadinstitute.org/cancer/software/genepattern/modules/docs/GSEA/14 GSEA v14] online documentation. | ||
| + | |||
| + | * Guide to [http://www.broadinstitute.org/gsea/doc/GSEAUserGuideFrame.html?_Interpreting_GSEA_Results interpreting GSEA Results] | ||
Revision as of 16:17, 14 January 2014
Contents
Overview
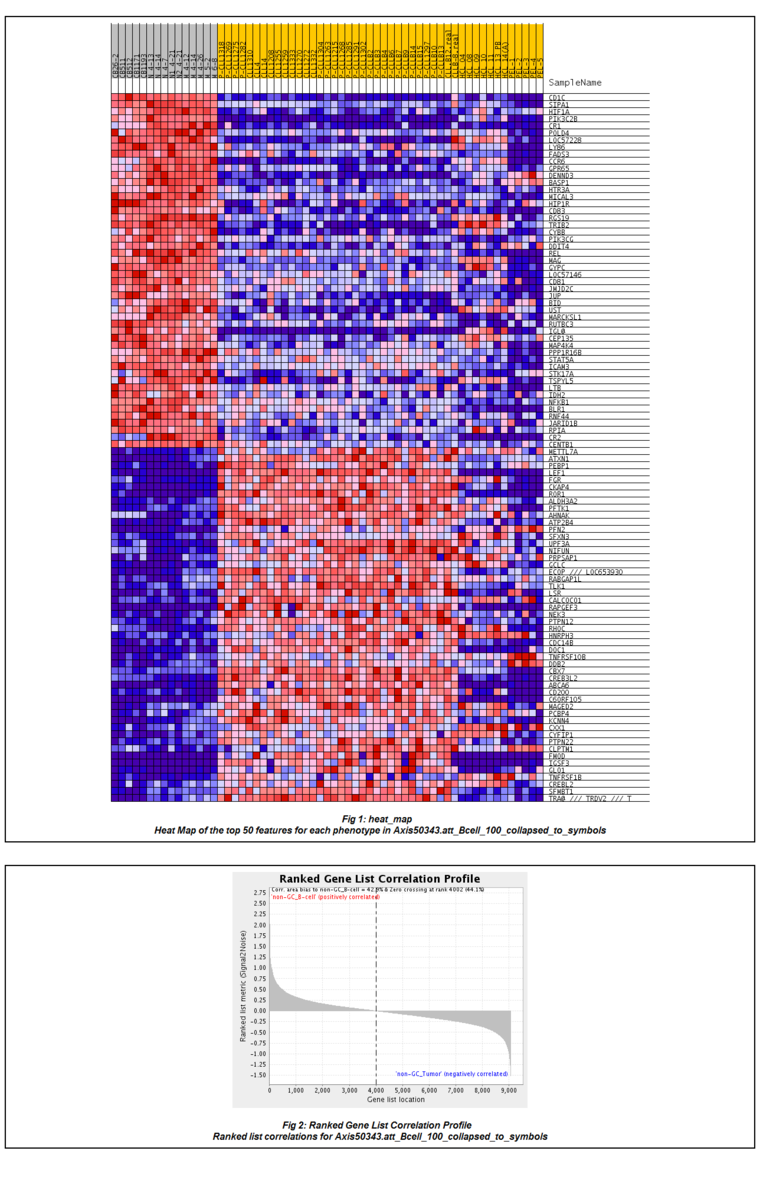
Gene Set Enrichment Analysis (Subramanian et al, 2005)
Prerequisites
- The "GSEA Analysis" and "GSEA Browser" components must be loaded in the Component Configuration Manager.
- An expression dataset must be loaded in the Workspace.
- Two (and only two) array sets must be activated in the Arrays component. They do not need to be marked "Case" or "Control", this will have no effect. These sets define the two classes used to calculate a measure of differential expression and from that the rank order of genes.
Parameters
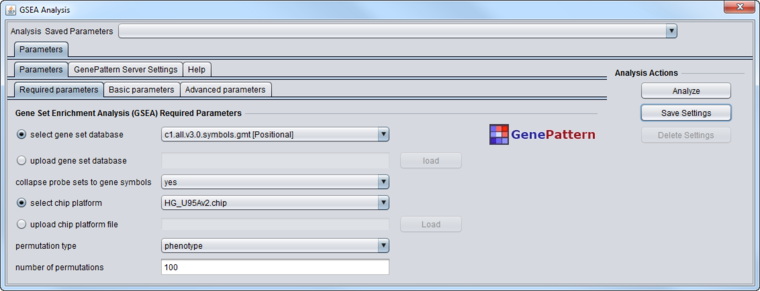
Required Parameters
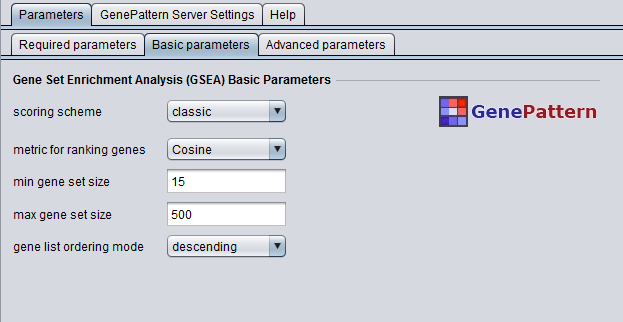
Basic Parameters
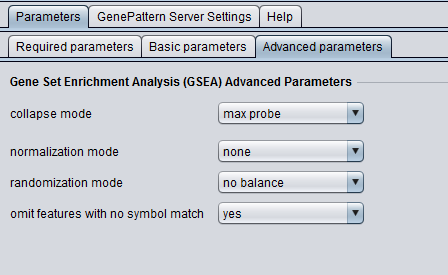
Advanced Parameters
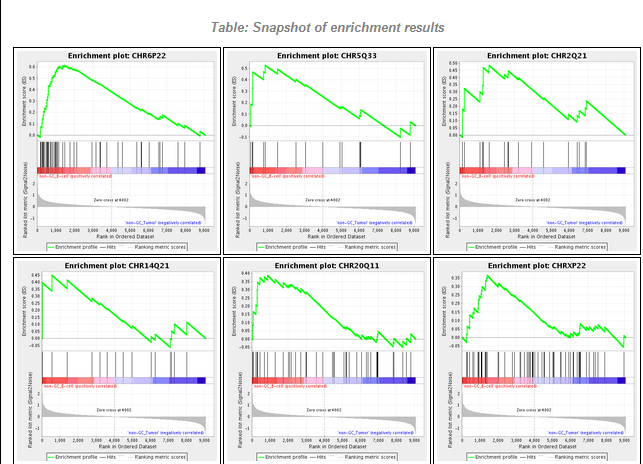
Results
References
- Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 102(43):15545-50. PubMed 16199517
- GenePattern modules documentation.
- GSEA v14 online documentation.
- Guide to interpreting GSEA Results