Difference between revisions of "Pattern Discovery"
(→Background) |
|||
| Line 8: | Line 8: | ||
The geWorkbench '''Pattern Discovery''' module uses an algorithm called '''SPLASH''' (Califano, 2000) to search for common patterns in sets of DNA or protein sequences. This type of search could be used, for example, to search for common regulatory elements in otherwise unrelated sequences. | The geWorkbench '''Pattern Discovery''' module uses an algorithm called '''SPLASH''' (Califano, 2000) to search for common patterns in sets of DNA or protein sequences. This type of search could be used, for example, to search for common regulatory elements in otherwise unrelated sequences. | ||
| − | For this tutorial, we will begin with the set of sequences retrieved as shown in the [[Tutorial_-_Sequence_Retrieval | Sequence | + | For this tutorial, we will begin with the set of 33 sequences retrieved as shown in the [[Tutorial_-_Sequence_Retrieval | Sequence Retrieval]] tutorial. These sequences derive from a cluster of genes showing similar expression pattern across a number of different experiments. |
('''Note''' - there currently is no provision for filtering out repeated sequences from genomic seqeuence. Results should be evaluated in this light). | ('''Note''' - there currently is no provision for filtering out repeated sequences from genomic seqeuence. Results should be evaluated in this light). | ||
Revision as of 17:34, 21 July 2006
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Background
The geWorkbench Pattern Discovery module uses an algorithm called SPLASH (Califano, 2000) to search for common patterns in sets of DNA or protein sequences. This type of search could be used, for example, to search for common regulatory elements in otherwise unrelated sequences.
For this tutorial, we will begin with the set of 33 sequences retrieved as shown in the Sequence Retrieval tutorial. These sequences derive from a cluster of genes showing similar expression pattern across a number of different experiments.
(Note - there currently is no provision for filtering out repeated sequences from genomic seqeuence. Results should be evaluated in this light).
Setting parameters and running
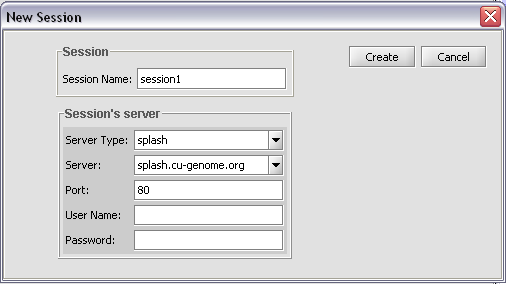
A number of parameters can be adjusted by the user, as shown in the figure, to adjust the sensitivity of the search. A user name must be entered, but it can be any name.
Push Create to start the search.
Viewing results
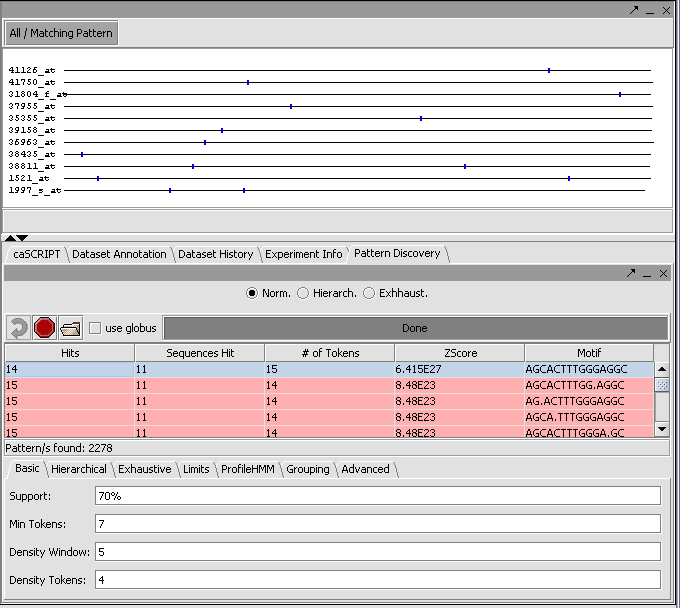
The result of the search can be viewed both in the Pattern Discovery module itself and in other sequence viewer modules such as "Sequence" and "Promoter".
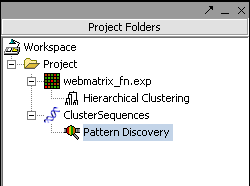
Results are added to the Projects Folder
The results of a run of Pattern Discovery are placed in the Project Folder:
References
Calfano, A. (2000). SPLASH: structural pattern localization analysis by sequential histograms. Bioinformatics, Apr;16(4):341-57.