Difference between revisions of "ARACNe"
(→Quick Example) |
|||
| Line 13: | Line 13: | ||
The resulting adjancency matrix will be displayed in the Cytoscape network visualization component. | The resulting adjancency matrix will be displayed in the Cytoscape network visualization component. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[Image:T_ARACNE_parameters.png]] | ||
| + | |||
| + | |||
| + | |||
| + | [[Image:T_ARACNE_parameters_List.png]] | ||
Revision as of 11:52, 16 October 2008
A new ARACNE tutorial is coming soon....
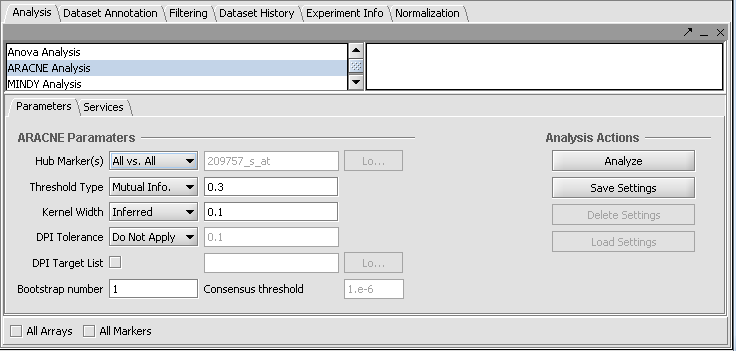
ARACNE in geWorkbench is used to determine interactions between a hub gene and a set of selected genes (or all genes in the set).
Quick Example
- Load a microarray dataset, e.g. Bcell-100.exp.
- Set a hub gene (e.g 1973_s_at)
- Click Analyze.
The resulting adjancency matrix will be displayed in the Cytoscape network visualization component.