Difference between revisions of "GenomeSpace"
(Created page with "==Overview== [http://www.genomespace.org/ GenomeSpace] allows for the transfer of data between a number of different genomics and bioinformatics software analysis platforms, incl...") |
(→Launch geWorkbench from the GenomeSpace Portal) |
||
| (53 intermediate revisions by the same user not shown) | |||
| Line 1: | Line 1: | ||
| + | {{TutorialsTopNav}} | ||
| + | |||
==Overview== | ==Overview== | ||
[http://www.genomespace.org/ GenomeSpace] allows for the transfer of data between a number of different genomics and bioinformatics software analysis platforms, including geWorkbench. GenomeSpace provides a cloud-based portal where data can be shared between these integrated platforms. geWorkbench includes a GenomeSpace component which allows direct upload and download of files to and from GenomeSpace. | [http://www.genomespace.org/ GenomeSpace] allows for the transfer of data between a number of different genomics and bioinformatics software analysis platforms, including geWorkbench. GenomeSpace provides a cloud-based portal where data can be shared between these integrated platforms. geWorkbench includes a GenomeSpace component which allows direct upload and download of files to and from GenomeSpace. | ||
A full description is available at [http://www.genomespace.org/what-is-genomespace What is GenomeSpace]. | A full description is available at [http://www.genomespace.org/what-is-genomespace What is GenomeSpace]. | ||
| + | |||
| + | The GenomeSpace site also provides a [http://www.genomespace.org/support/guides/tool-guide/sections/geworkbench-GS-capabilities detailed tutorial] on the use of geWorkbench with GenomeSpace. | ||
==Shareable file formats== | ==Shareable file formats== | ||
| − | The GenomeSpace | + | GenomeSpace acts as a neutral file transfer host. It includes a selection of file format conversion routines which can be selected by the user. The conversion can be performed directly on the web portal or within the GenomeSpace component in geWorkbench. Currently, GenomeSpace can convert GenePattern GCT files to geWorkbench EXP format files. The reverse conversion is not available at this time, however EXP files can be uploaded from geWorkbench into GenomeSpace. |
| + | |||
| + | ==The GenomeSpace Portal== | ||
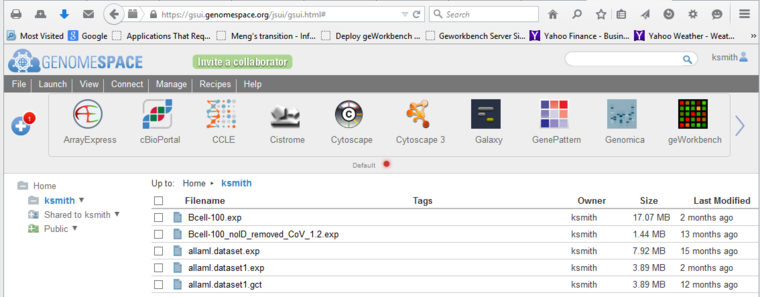
| + | Files can be uploaded to the GenomeSpace portal directly or from enabled programs. The screenshot below shows the cloud-based portal, to which a GCT format file has been uploaded. | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_Portal.png|{{ImageMaxWidth}}]] | ||
| + | |||
| + | |||
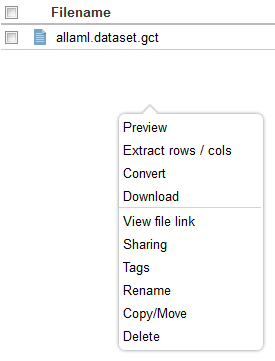
| + | Right-clicking on an uploaded file provides a menu of actions that can be performed, including format conversion for known file types. | ||
| + | |||
| + | [[Image:GenomeSpace_Portal_Options.png]] | ||
| + | |||
| + | ==GenomeSpace in geWorkbench== | ||
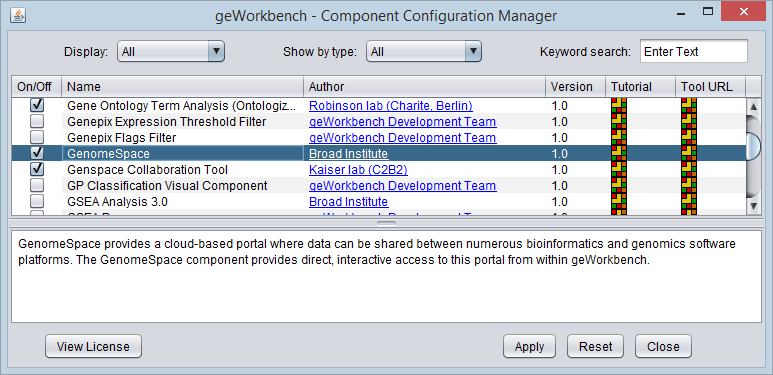
| + | The GenomeSpace component is now loaded by default in the [[Component_Configuration_Manager|Component Configuration Manager (CCM)]]. If can be unloaded/loaded by checking/unchecking the adjacent box and hitting "Apply" and then "Close". | ||
| + | |||
| + | [[Image:GenomeSpace_CCM.png]] | ||
| + | |||
| + | |||
| + | The GenomeSpace component appears in the Visual area of geWorkbench. | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_Empty.png]] | ||
| + | |||
| + | |||
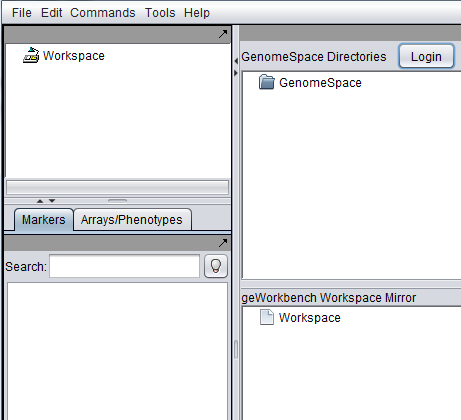
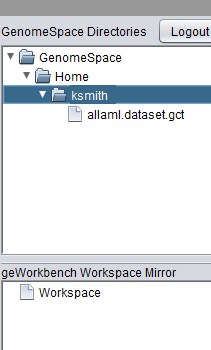
| + | The GenomeSpace component has two panes: | ||
| + | * '''GenomeSpace Directories''' - The upper pane reflects the data on the cloud-based portal. | ||
| + | * '''geWorkbench Workspace Mirror''' - The lower pane mirrors data that is loaded in the geWorkbench Workspace (shown at upper left in the figure). | ||
| + | |||
| + | |||
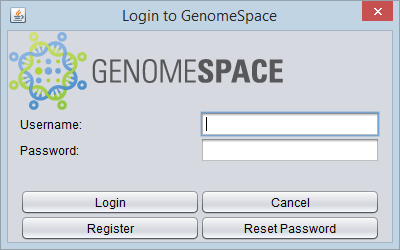
| + | To share data, the user must log in within the component in geWorkbench by pushing the "Login" button. An existing GenomeSpace account can be used, or a new account created. | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_Login.png]] | ||
| + | |||
| + | |||
| + | Once logged in, the shared workspace is displayed in geWorkbench, along with any data files that may already be present there. | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_data1.png]] | ||
| + | |||
| + | ==GenomeSpace Directories== | ||
| + | ===File Operations Overview=== | ||
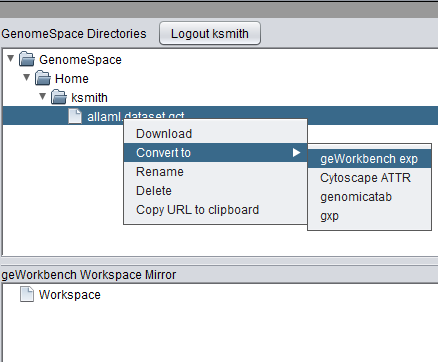
| + | Right-clicking on a data file in the upper pane (GenomeSpace Directories) will provide a number of actions available for that file: | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_Convert_Menu.png]] | ||
| + | |||
| + | |||
| + | * '''Download''' - Download the file into geWorkbench. | ||
| + | * '''Convert''' - Convert the file to the selected format. "geWorkbench exp" is the format that can be used in geWorkbench. | ||
| + | * '''Rename''' - Rename the file. | ||
| + | * '''Delete''' - Delete the file. | ||
| + | * '''Copy URL to clipboard''' - Copy the URL of the selected folder or data file on the GenomeSpace server to the clipboard. | ||
| + | |||
| + | ===Convert=== | ||
| + | ====Available Converters==== | ||
| + | The GenomeSpace site provides full documentation on [http://www.genomespace.org/support/guides/user-guide/sections/converting-files supported file format conversions]. | ||
| + | |||
| + | Available converters for geWorkbench file formats: | ||
| + | |||
| + | * GCT to EXP. EXP is a geWorkbench gene expression format. | ||
| + | * ADJ to XGMML. ADJ is a gene network format used by [[ARACNe]]. | ||
| + | |||
| + | ====GCT to EXP Conversion==== | ||
| + | GCT files in GenomeSpace can be converted to geWorkbench EXP files either manually or automatically. | ||
| + | * If a GCT file is selected for download into geWorkbench, it will automatically be converted to an EXP file. | ||
| + | * A GCT file can also be converted to an EXP file by right-clicking on it and selecting "Convert to->geWorkbench exp". | ||
| + | * The conversion can also be directly performed in the GenomeSpace web portal. | ||
| + | * '''Note''' - If the file is converted from GCT to EXP directly on the GenomeSpace portal, the GCT file is retained. However, if the conversion is done in the GenomeSpace component in geWorkbench, the original GCT file is replaced with the EXP file. | ||
| + | |||
| + | ===Download=== | ||
| + | When Download is selected, the selected file (currently only download in EXP format is supported) will be downloaded from the portal into geWorkbench. | ||
| + | |||
| + | During the download process, the standard geWorkbench expression file loading dialog will open and allow the user to add an annotation file. | ||
| + | |||
| + | On successful download, the file will appear both in the geWorkbench Workspace, and in the mirror of the Workspace which is shown in the lower pane of the GenomeSpace component. | ||
| + | |||
| + | * At this time, only expression formats GCT and EXP can be downloaded into geWorkbench. | ||
| + | * ADJ files cannot be downloaded into geWorkbench. | ||
| + | |||
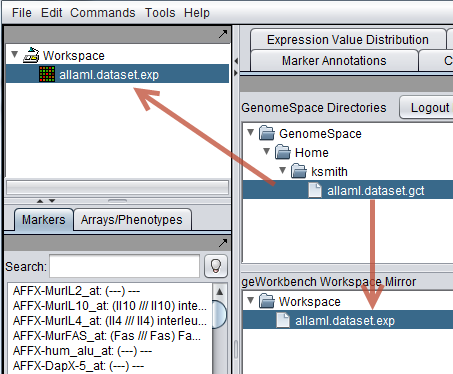
| + | This example shows the result of automatic conversion from GCT to EXP format during download. | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_Downloaded_File.png]] | ||
| + | |||
| + | ===Workspace Folder Operations=== | ||
| + | |||
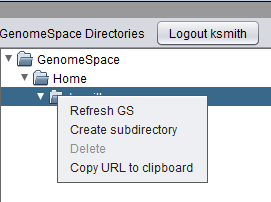
| + | Right-clicking on any folder in the GenomeSpace Workspace will provide a menu of options. | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_Folder_Menus.png]] | ||
| + | |||
| + | |||
| + | * '''Refresh GS''' - Refresh the GenomeSpace workspace to reflect any remote changes. | ||
| + | * '''Create subdirectory''' - Create a new subdirectory in the Workspace. | ||
| + | * '''Delete''' - Only available for files. Deletes the file from the GenomeSpace Workspace. | ||
| + | * '''Copy URL to clipboard''' - Copy the URL of the selected folder or data file on the GenomeSpace server to the clipboard. | ||
| + | |||
| + | ==geWorkbench Workspace Mirror== | ||
| + | ===Upload=== | ||
| + | |||
| + | * Currently, only EXP format expression files can be uploaded. | ||
| + | * ADJ files cannot be uploaded. | ||
| + | |||
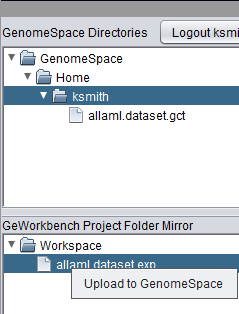
| + | Right-clicking on a file will bring up an upload button, to upload the file to the GenomeSpace Workspace. | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_Upload_Menu.png]] | ||
| + | |||
| + | |||
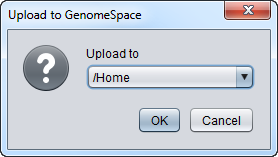
| + | After pushing the Upload button, the user will be prompted for a destination directory in the GenomeSpace Workspace. | ||
| + | |||
| + | [[Image:GenomeSpace_Upload_Location1.png]] | ||
| + | |||
| + | |||
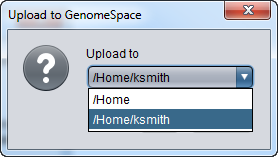
| + | A pulldown menu in the upload dialog offers the list of available directories. | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_Upload_Location2.png]] | ||
| + | |||
| + | |||
| + | The figure below shows that the EXP file has been uploaded from geWorkbench to GenomeSpace. | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_Uploaded_File.png]] | ||
| + | |||
| + | ==Launch geWorkbench from the GenomeSpace Portal== | ||
| + | geWorkbench can be launched directly from the GenomeSpace Portal, using Java Web Start. If geWorkbench is not already installed, you will be prompted to download and install it. | ||
| + | |||
| + | Note - due to the complexity of the below steps, you can also just install or launch geWorkbench independtly of GenomeSpace. As soon as you start geWorkbench, the GenomeSpace component will be available for logging in. | ||
| + | |||
| + | Because Java is subject to increasingly stringent security, you may need to carry out the following steps. | ||
| + | |||
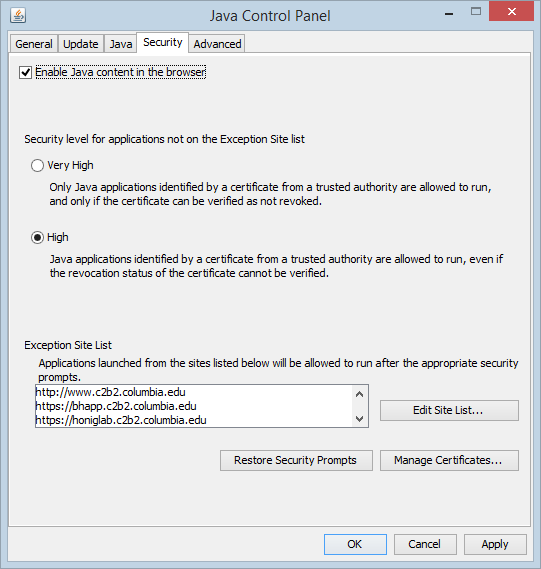
| + | You must have a Java JRE installed on your computer already. In your Java Control Panel (in Windows Control Panel, in Mac OS X System Preferences), go to the Security tab and add an exception for "http://www.c2b2.columbia.edu". | ||
| + | |||
| + | |||
| + | On Windows, this can be done by going to Control Panel -> Java -> Security. On the Mac, go to System Preferences -> Java -> Security. | ||
| + | |||
| + | Once in the Java Security tab, two steps are needed: | ||
| + | |||
| + | * Check the box "Enable Java content in the browser" | ||
| + | *Add an exception to allow Java to run for a particular URL. | ||
| + | ** Click "Edit Site List" and then "Add". | ||
| + | ** Type in the URL for www.columbia.edu. You can also add the other two for the [[MarkUs]] component if you wish: | ||
| + | *** http://www.c2b2.columbia.edu | ||
| + | *** https://bhapp.c2b2.columbia.edu. | ||
| + | *** https://honiglab.c2b2.columbia.edu. | ||
| + | |||
| + | *Then click the "OK" button. | ||
| + | |||
| + | |||
| + | |||
| + | [[Image:Java_Control_Panel.png]] | ||
| + | |||
| + | |||
| + | Clicking on the geWorkbench icon in GenomeSpace will launch the Java Web Start application: | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_launchHelper.png]] | ||
| + | |||
| + | |||
| + | Check the box for "I accept the risk..." | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_SecurityWarning.png]] | ||
| + | |||
| + | |||
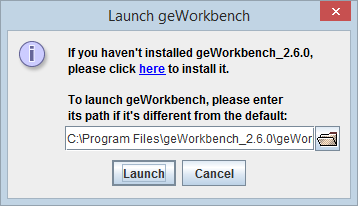
| + | A dialog box will appear to confirm the installation directory for geWorkbench. If geWorkbench was installed in another location, please enter that into the box. | ||
| + | |||
| + | |||
| + | [[Image:GenomeSpace_Install_geWorkbench.png]] | ||
Latest revision as of 18:33, 6 March 2015
Contents
Overview
GenomeSpace allows for the transfer of data between a number of different genomics and bioinformatics software analysis platforms, including geWorkbench. GenomeSpace provides a cloud-based portal where data can be shared between these integrated platforms. geWorkbench includes a GenomeSpace component which allows direct upload and download of files to and from GenomeSpace.
A full description is available at What is GenomeSpace.
The GenomeSpace site also provides a detailed tutorial on the use of geWorkbench with GenomeSpace.
GenomeSpace acts as a neutral file transfer host. It includes a selection of file format conversion routines which can be selected by the user. The conversion can be performed directly on the web portal or within the GenomeSpace component in geWorkbench. Currently, GenomeSpace can convert GenePattern GCT files to geWorkbench EXP format files. The reverse conversion is not available at this time, however EXP files can be uploaded from geWorkbench into GenomeSpace.
The GenomeSpace Portal
Files can be uploaded to the GenomeSpace portal directly or from enabled programs. The screenshot below shows the cloud-based portal, to which a GCT format file has been uploaded.
Right-clicking on an uploaded file provides a menu of actions that can be performed, including format conversion for known file types.
GenomeSpace in geWorkbench
The GenomeSpace component is now loaded by default in the Component Configuration Manager (CCM). If can be unloaded/loaded by checking/unchecking the adjacent box and hitting "Apply" and then "Close".
The GenomeSpace component appears in the Visual area of geWorkbench.
The GenomeSpace component has two panes:
- GenomeSpace Directories - The upper pane reflects the data on the cloud-based portal.
- geWorkbench Workspace Mirror - The lower pane mirrors data that is loaded in the geWorkbench Workspace (shown at upper left in the figure).
To share data, the user must log in within the component in geWorkbench by pushing the "Login" button. An existing GenomeSpace account can be used, or a new account created.
Once logged in, the shared workspace is displayed in geWorkbench, along with any data files that may already be present there.
GenomeSpace Directories
File Operations Overview
Right-clicking on a data file in the upper pane (GenomeSpace Directories) will provide a number of actions available for that file:
- Download - Download the file into geWorkbench.
- Convert - Convert the file to the selected format. "geWorkbench exp" is the format that can be used in geWorkbench.
- Rename - Rename the file.
- Delete - Delete the file.
- Copy URL to clipboard - Copy the URL of the selected folder or data file on the GenomeSpace server to the clipboard.
Convert
Available Converters
The GenomeSpace site provides full documentation on supported file format conversions.
Available converters for geWorkbench file formats:
- GCT to EXP. EXP is a geWorkbench gene expression format.
- ADJ to XGMML. ADJ is a gene network format used by ARACNe.
GCT to EXP Conversion
GCT files in GenomeSpace can be converted to geWorkbench EXP files either manually or automatically.
- If a GCT file is selected for download into geWorkbench, it will automatically be converted to an EXP file.
- A GCT file can also be converted to an EXP file by right-clicking on it and selecting "Convert to->geWorkbench exp".
- The conversion can also be directly performed in the GenomeSpace web portal.
- Note - If the file is converted from GCT to EXP directly on the GenomeSpace portal, the GCT file is retained. However, if the conversion is done in the GenomeSpace component in geWorkbench, the original GCT file is replaced with the EXP file.
Download
When Download is selected, the selected file (currently only download in EXP format is supported) will be downloaded from the portal into geWorkbench.
During the download process, the standard geWorkbench expression file loading dialog will open and allow the user to add an annotation file.
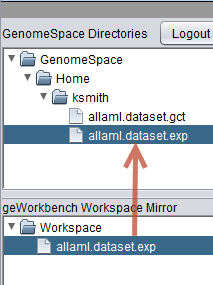
On successful download, the file will appear both in the geWorkbench Workspace, and in the mirror of the Workspace which is shown in the lower pane of the GenomeSpace component.
- At this time, only expression formats GCT and EXP can be downloaded into geWorkbench.
- ADJ files cannot be downloaded into geWorkbench.
This example shows the result of automatic conversion from GCT to EXP format during download.
Workspace Folder Operations
Right-clicking on any folder in the GenomeSpace Workspace will provide a menu of options.
- Refresh GS - Refresh the GenomeSpace workspace to reflect any remote changes.
- Create subdirectory - Create a new subdirectory in the Workspace.
- Delete - Only available for files. Deletes the file from the GenomeSpace Workspace.
- Copy URL to clipboard - Copy the URL of the selected folder or data file on the GenomeSpace server to the clipboard.
geWorkbench Workspace Mirror
Upload
- Currently, only EXP format expression files can be uploaded.
- ADJ files cannot be uploaded.
Right-clicking on a file will bring up an upload button, to upload the file to the GenomeSpace Workspace.
After pushing the Upload button, the user will be prompted for a destination directory in the GenomeSpace Workspace.
A pulldown menu in the upload dialog offers the list of available directories.
The figure below shows that the EXP file has been uploaded from geWorkbench to GenomeSpace.
Launch geWorkbench from the GenomeSpace Portal
geWorkbench can be launched directly from the GenomeSpace Portal, using Java Web Start. If geWorkbench is not already installed, you will be prompted to download and install it.
Note - due to the complexity of the below steps, you can also just install or launch geWorkbench independtly of GenomeSpace. As soon as you start geWorkbench, the GenomeSpace component will be available for logging in.
Because Java is subject to increasingly stringent security, you may need to carry out the following steps.
You must have a Java JRE installed on your computer already. In your Java Control Panel (in Windows Control Panel, in Mac OS X System Preferences), go to the Security tab and add an exception for "http://www.c2b2.columbia.edu".
On Windows, this can be done by going to Control Panel -> Java -> Security. On the Mac, go to System Preferences -> Java -> Security.
Once in the Java Security tab, two steps are needed:
- Check the box "Enable Java content in the browser"
- Add an exception to allow Java to run for a particular URL.
- Click "Edit Site List" and then "Add".
- Type in the URL for www.columbia.edu. You can also add the other two for the MarkUs component if you wish:
- Then click the "OK" button.
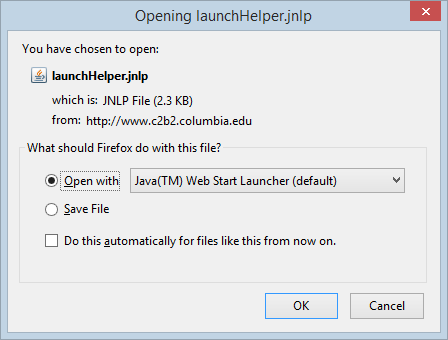
Clicking on the geWorkbench icon in GenomeSpace will launch the Java Web Start application:
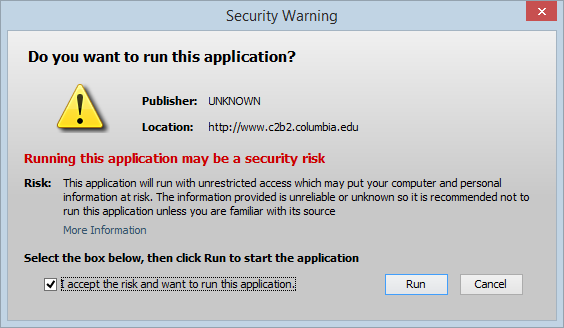
Check the box for "I accept the risk..."
A dialog box will appear to confirm the installation directory for geWorkbench. If geWorkbench was installed in another location, please enter that into the box.