Difference between revisions of "Jmol"
| Line 30: | Line 30: | ||
#. The molecular structure will be displayed in the Jmol component. | #. The molecular structure will be displayed in the Jmol component. | ||
| − | ==Jmol | + | |
| − | Jmol has a number of optional settings to modify the display. | + | ==Jmol Display Options== |
| + | |||
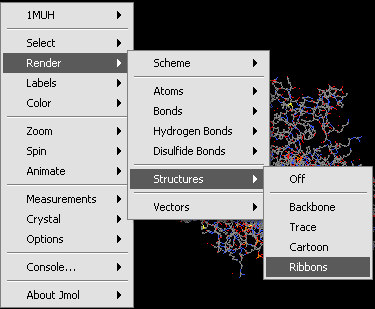
| + | Jmol has a number of optional settings to modify the display. These include operations to select, render, color and label the molecule and its constituent parts. As an example, the rendering sub-menu is depicted below: | ||
| + | |||
| + | [[Image:T_JMOL_RenderOption.png]] | ||
| + | |||
| + | Jmol Rendering submenu. | ||
Revision as of 18:04, 21 December 2006
Outline
This tutorial contains
- . an overview of Jmol and
- . instructions on how to view molecular structure (PDB format) files within geWorkbench.
Overview
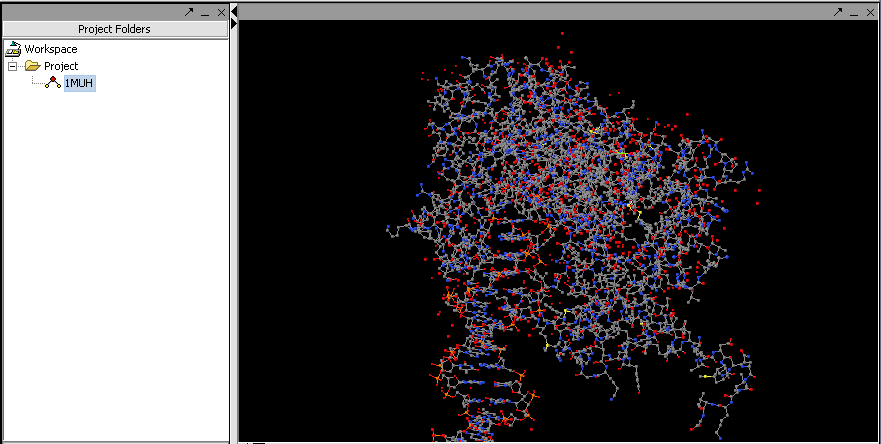
Jmol is a molecular structure viewer. It is from an open-source project which can be found at jmol.sourceforge.net. It is used in geWorkbench to view PDB format files. In the figure below, it has been used to open the file 1muh.pdb, available from the Protein Data bank at PDB:1muh
The file 1muh.pdb displayed in Jmol.
Loading a PDB file
- . Create a new project or select an existing project in the Project Folders component.
- . Right-click on the project to bring up the Open File dialog.
- . Select the file type as "PDB File Format".
- . Navigate using the file browswer to the desired PDB format file and read it in. (these files end with a ".pdb" filename suffix).
- . The molecular structure will be displayed in the Jmol component.
Jmol Display Options
Jmol has a number of optional settings to modify the display. These include operations to select, render, color and label the molecule and its constituent parts. As an example, the rendering sub-menu is depicted below:
Jmol Rendering submenu.