Difference between revisions of "Pudge"
(→Parameters and running Pudge) |
(→Overview) |
||
| Line 5: | Line 5: | ||
The Pudge component serves as a front end to the web-based Pudge protein structure modeling service developed by the Honig lab at Columbia University. | The Pudge component serves as a front end to the web-based Pudge protein structure modeling service developed by the Honig lab at Columbia University. | ||
| − | Computational protein structure prediction using sequence homology is a process | + | Computational protein structure prediction using sequence homology is a process that exploits the observation that proteins with similar sequences usually have similar structures as well. While this idea is simple in theory, in practice, the process of using sequence homology to predict protein structure is intricate and complex. The process is generally divided into five stages, involving the use of a combination of techniques from a variety of scientific disciplines. See Petrey & Honig, 2005 for a description of the overall protein structure prediction process. |
| − | |||
==Parameters and running Pudge== | ==Parameters and running Pudge== | ||
Revision as of 12:51, 3 December 2009
Overview
The Pudge component serves as a front end to the web-based Pudge protein structure modeling service developed by the Honig lab at Columbia University.
Computational protein structure prediction using sequence homology is a process that exploits the observation that proteins with similar sequences usually have similar structures as well. While this idea is simple in theory, in practice, the process of using sequence homology to predict protein structure is intricate and complex. The process is generally divided into five stages, involving the use of a combination of techniques from a variety of scientific disciplines. See Petrey & Honig, 2005 for a description of the overall protein structure prediction process.
Parameters and running Pudge
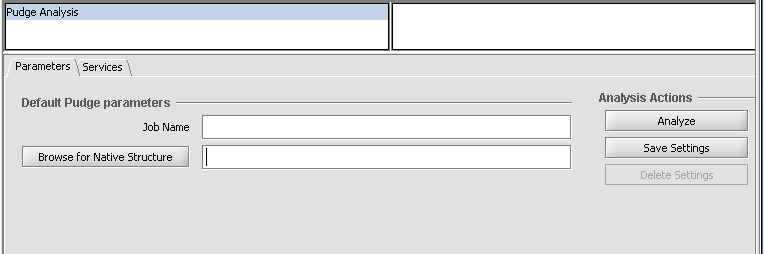
If Pudge has been loaded via the CCM, it will appear in the Analysis area when a FASTA sequence file is loaded and selected in the Project Folders area.
To run Pudge,
1. add a job name in the Job Name text field.
2. optionally, add a PDB protein structure file to serve as a starting structural template.
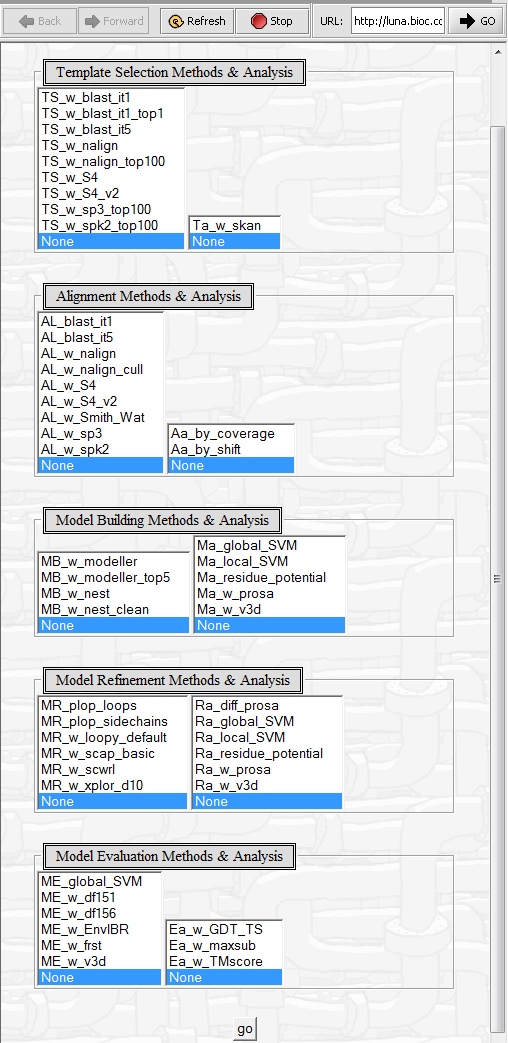
3. click Analyze. This will cause a web browser page to be displayed which contains a large number of additional configuration options.
4. Once any changes have been made, click on Go to launch the job.
Results will be displayed in a web browser window within geWorkbench. From there, once can save result files in PDB format and load them into geWorkbench for viewing and further manipulation.