Difference between revisions of "Marker Annotations"
(→New Content) |
|||
| Line 57: | Line 57: | ||
==New Content== | ==New Content== | ||
| + | |||
| + | Marker Annotations | ||
| + | The Marker Annotations component enables the retrieval of biological annotation information for a collection of genes. For every gene, the following data can be retrieved: | ||
| + | |||
| + | * A set of pathways containing the gene. | ||
| + | * A set of gene-disease and gene-compound associations derived from the literature articles. | ||
| + | |||
| + | All annotations are retrieved from remote servers maintained by the National Cancer Institute (NCI). The data in those server come from the following sources: | ||
| + | |||
| + | * Pathways: NCI's Pathway Interaction Database (PID). PID pathways come from 3 sources: BioCarta, Reactome and "NCI-Nature Curated". Information about the PID and each of the contributing sources is available at: http://pid.nci.nih.gov/userguide/database_content.shtml. These pathways are stored in servers used by the Cancer Gene Anatomy Project (CGAP, http://cgap.nci.nih.gov/). | ||
| + | * Gene-disease/compound associations: the Cancer Gene Index (CGI) data base. The reported assocations are extracted from article abstracts using a combination of automatic text mining, semi-automatic verification, and manual curation. Project details are available at: http://ncicb.nci.nih.gov/NCICB/projects/cgdcp. | ||
| + | |||
| + | Submit Query | ||
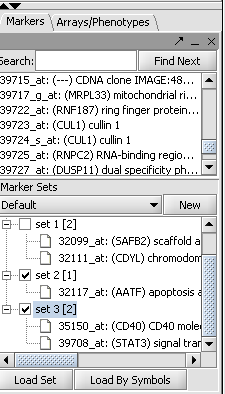
| + | The Marker Annotations module will retrieve information for all markers that belong to activated marker sets, or, more precisely, for the genes corresponding to those markers: | ||
| + | |||
| + | [[Image:Select_marker_sets.png]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | E.g., in the example shown above, information will be retrieved about the genes AATF, CD40, STAT3. Checkboxes at the bottom of the component's user interface can be used to specify which data source(s) to query: CGAP, CGI or both. For CGAP, the associated drop-down can be used to designate the target organism for which annotations are retrieved: human (the default) or mouse. Clicking the "Retrieve Annotations" button initiates the communication with the NCI servers: | ||
| + | |||
| + | |||
| + | |||
| + | [[Image:Data_source_checkboxes.png]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Pathway and Gene Annotations | ||
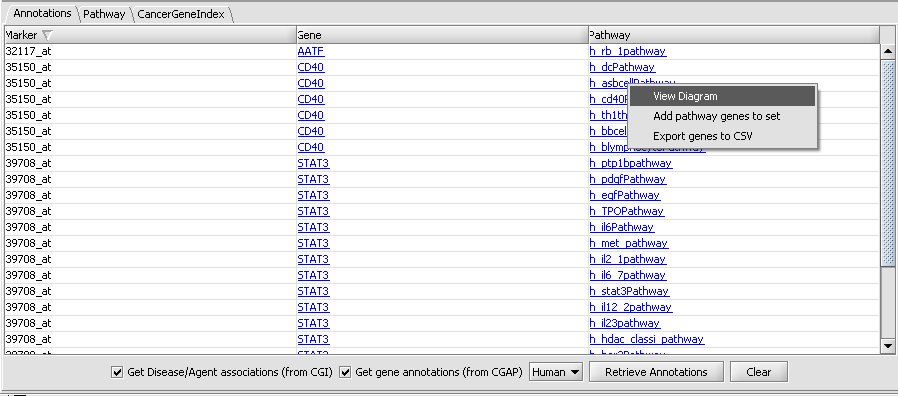
| + | The "Annotations" tab presents a summary listing of the annotations retrieved from CGAP: | ||
| + | |||
| + | [[Image:CGAP_summary_page.png]] | ||
| + | |||
| + | |||
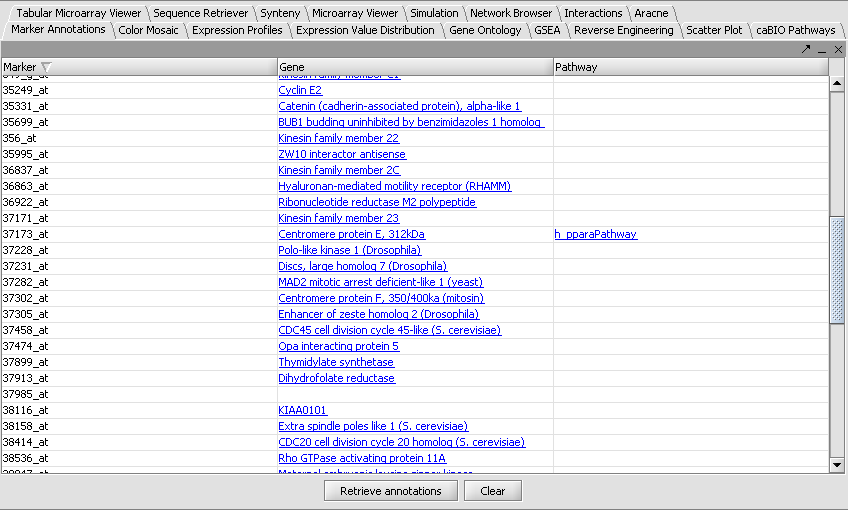
| + | The listing contains at least one row for each gene which annotation infromation is available for. If a gene is associated with more than one pathways, then one row for every pathway is listed (e.g., as is the case above for CD40 and STAT3). Every row displays the marker (i.e., probeset) id, the corresponding gene name and the name of the associated pathway. Clicking on a pathway brings up a popup menu offering a number options: | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
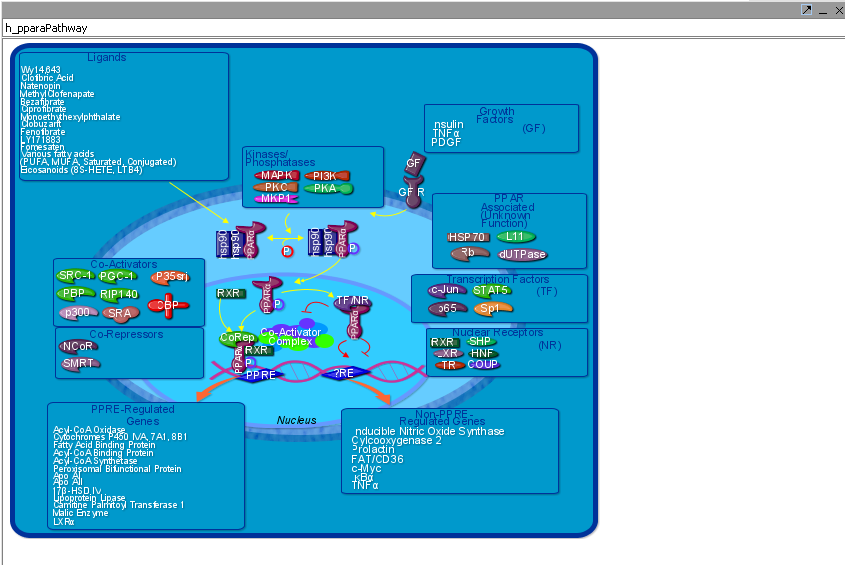
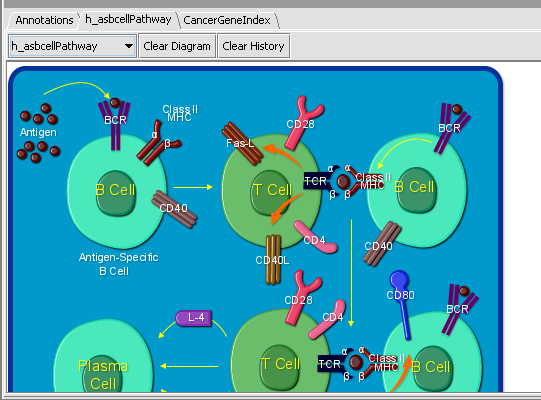
| + | * View Diagram: available only for BioCarta pathways. Such pathways are accompanied by images offering a graphical/artistic rendition of the pathway. Selecting the "View Diagram" option will display this image wihtin the "Pathway" tab. | ||
| + | * Add pathway genes to set: extracts the pathway genes for which there are associated probes in the microrarray set currently selected by the user and places all such probes in a new marker set within the "Markers" component (by default, the marker set is named after the pathway). | ||
| + | * Export genes to CSV: creates a new text file containing a listing of all pathway genes. The file format (csv = comma separated values) is compatible with Microsoft Excel. | ||
| + | |||
[[Image:Biocarta.png]] | [[Image:Biocarta.png]] | ||
| + | |||
| + | |||
| + | A BioCarta pathway image is displayed above after selecting the "View Dagram" option from the "Annotations" tab. The drop-down box, on the top left corner above the diagram, shows the name of the currently displayed diagram. The component keeps a history of all BioCarta diagrams selected by the user; using the drop-down it is possible to switch among the correponding pathway images. The "Clear Diagram" button clears the currently displayed diagram. The "Clear History" button both clears the currently displayed diagram and removes all pathway history information from the pathway name drop-down box. | ||
| + | |||
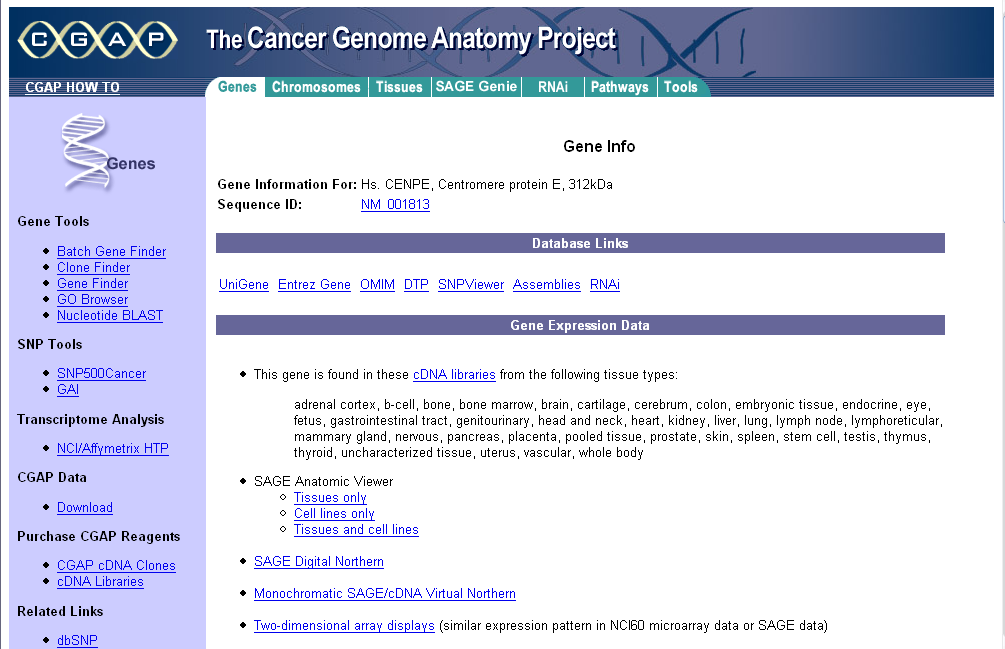
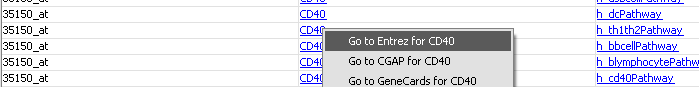
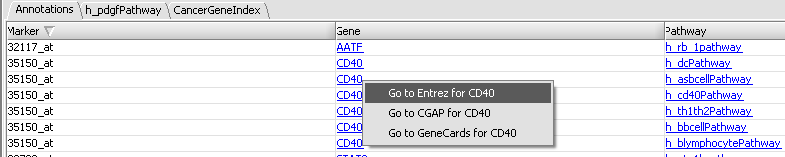
| + | In the "Annotations" tab, it is also possible to click on a gene name and explore functional annotation information from a number of sources (Entrez, CGAP, GeneCards): | ||
[[Image:CGAP_click_on_gene.png]] | [[Image:CGAP_click_on_gene.png]] | ||
| − | [[Image: | + | |
| + | Cancer Gene Index | ||
| + | For many genes, there are hundreds of records in the CGI database. Retrieving all those records at once can be a very time consuming operation, especially if the query invlolves many genes. To avoid very long waits, the retrieval of the data occurs in 2 stages. In the first stage, at most 10 records for each association type (gene-disease/gene-compound) are being fetched (for each query gene). Data retrieved as displayed in the CancerGeneIndex tab: | ||
| + | |||
| + | |||
| + | [[Image:CGI_summary_page.png]] | ||
| + | |||
| + | |||
| + | The user interface is divided in two regions, sharing the same overall structure and functionality: the table on the left displays gene-disease associations, while the table on the right is used for reporting gene-compound associations. To avoid redundancy, the paragraphs that follow describe only the gene-disease table. The exact same description applies in the case of the gene-compound table. | ||
| + | |||
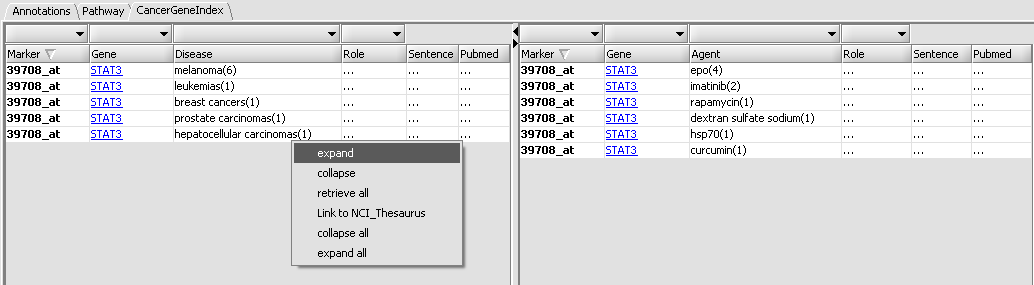
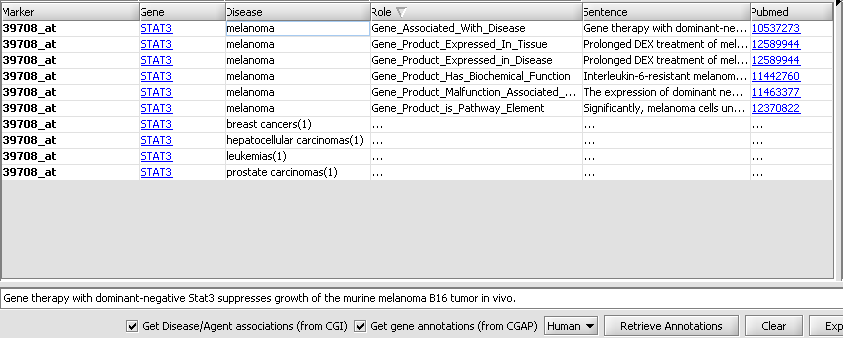
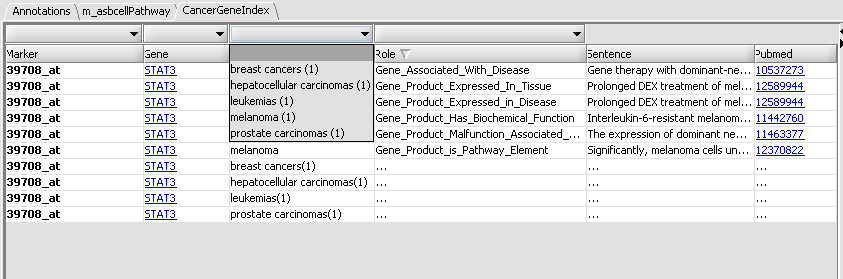
| + | Each table row represent an association between a query gene and a disease. The first column contains the name of the probeset associated with the query gene. For any given gene, if its corresponding probeset name appears in bold-face, this means that there are more gene-disease records on the CGI server that have yet to be fetched (beyond the 10 records acquired in the initial retrieval stage). The disease name within a row is followed by a number in parentheses. This number indicates how many records (among those fetched) support the reported association. E.g, in the image above, the first row indicates that there are 6 distinct records linking STAT3 to melanoma. A detailed listing of these 6 records is available by "expanding" the row. This can be achieved by right-clicking on the row and selecting "expand" from the ensuing popup menu: | ||
| + | |||
[[Image:CGI_expanded.png]] | [[Image:CGI_expanded.png]] | ||
| − | |||
| − | [[Image: | + | Each detailed record contains 3 additional pieces of information: |
| + | |||
| + | * Role: a curator-assigned description of the kind of association being reported. The values in this column come from a controlled vocabulary (developed to support the CGI database creation effort). | ||
| + | * Sentence: the actual article abstract sentence used to derive the reported gene-disease association. The full sentence is displayed in the text area at the bottom portion of the interface (it is also availalbe as a tooltip text, by mousing over the "Sentence" column). | ||
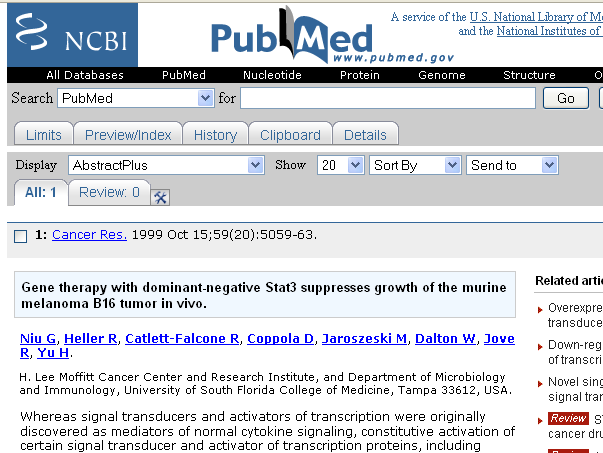
| + | * Pubmed: the Pubmed ID of the source article. Clicking on the Pubmed link brings up in the web browser the corresponding Pubmed abstract page (in this example, the sentence used for deriving the gene-disease association is actually the paper title): | ||
| + | |||
| + | |||
| + | [[Image:Pubmed.png]] | ||
| + | |||
| + | |||
| + | |||
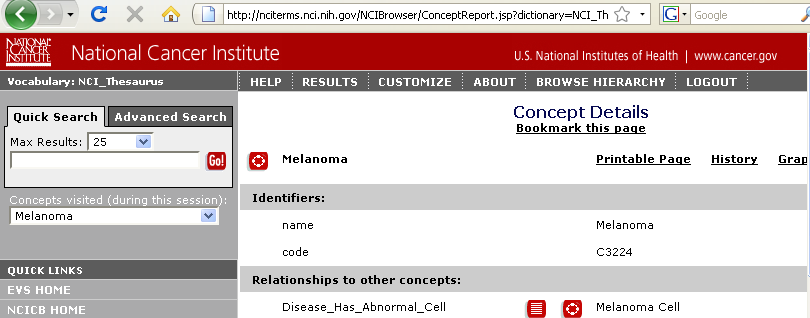
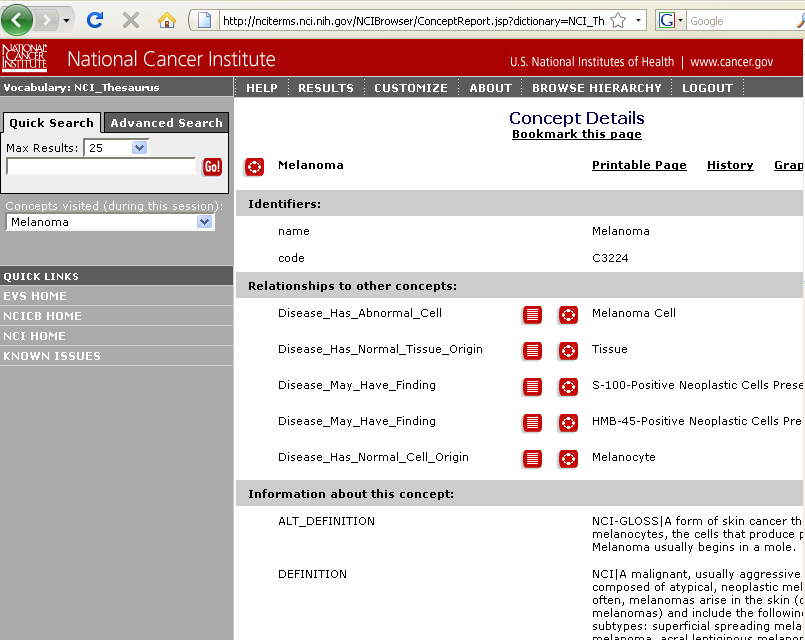
| + | It is also possible to link out to the NCI thesaurus in order to see the definition of the disease being associated with a gene; this is achieved by right-clicking on the table row for the gene-disease association and selecting the "Link to NCI_Thesaurus" option from the popup: | ||
| + | |||
| + | [[Image:NCI_thesaurus_disease_small.png]] | ||
| + | |||
| + | |||
| + | |||
| + | It should also be noted that the user interface allows ordering and filtering of the data (the latter can be very useful if there are many records being displayed): | ||
| + | |||
| + | * Ordering: the table rows can be sorted alphabetically by the contents of any column, by clicking on a column heading. | ||
| + | * Filtering: the drop down boxes that appear above the columns "Marker", "Gene", "Disease", and "Role" contain one value for each distinct entry within those columns. They can be used to select only records that contain the designated values. Of note is the drop-down associated with the "Disease" column: | ||
[[Image:CGI_disease_dropdown.png]] | [[Image:CGI_disease_dropdown.png]] | ||
| − | |||
| − | |||
| − | [[Image: | + | The parantheses next to a disease name indicate how many distinct genes (among those included in the user query) are associated with this particular disease. |
| + | |||
| + | It should be noted that these numbers are calculated using only fetched records. As mentioned above, the first stage of the infromation retrieval will fetch at most 10 records per gene. The remaining records asscociated with a gene can be retrieved from the CGI server by right-clicking on a table row corresponding to the gene and by selecting the popup menu option "retrieve" all: | ||
| + | |||
| + | ****IMAGE MISSING HERE**** | ||
| + | |||
| + | After all gene-disease association records for a given gene have been fetched, the bold-face type of its associated probeset name is removed. | ||
| + | |||
| + | Finally,by clicking on the "Export" button at the bottom of the user interfce, the contents of the gene-disease and the gene-compound tables displayed within the CancerGeneIndex tab can be exported as comma separated values text files for further analysis or/and visualization by spreadsheet software. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Extra1 | ||
| + | |||
| + | [[Image:CGAP_summary_page_gene.png]] | ||
| + | |||
| + | Extra2 | ||
| + | |||
| + | [[Image:CGI_expanded.png]] | ||
| − | |||
| − | + | Extra3 | |
| − | |||
| − | [[Image: | + | [[Image:NCI_thesaurus_disease_big.png]] |
Revision as of 00:14, 4 August 2009
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Outline
In this tutorial, we will:
- . Load a set of markers into the Markers component.
- . Retrieve annotations from NCI's CGAP database.
- . View a gene annotation page.
- . View a BioCarta pathway diagram in the caBIO Pathways viewer.
Overview
The Marker Annotations component retrieves two types of information for a group of genes. It retrieves links to pages from the CGAP database at NCI containing textual information. It also contains links to BioCarta pathway diagrams provided through NCI's caBIO data service. For this tutorial, we will examine a group of genes selected in the Hierarchical Clustering tutorial.
Loading a set of markers
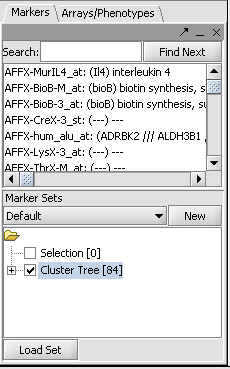
You must select and activate a group of markers (genes) in the Markers component. You can use for example the 84 markers selected in the hierarchical clustering example. However, as it takes about 5 seconds to retrieve the annotations for each marker, you can also load or select a smaller set of markers.
- To load a previously saved set of markers, go to the Markers component and click the "Load Set" button.
- The set previously obtained via hierarchical clustering is in the tutorial data file "cluster_tree_total_pearsons_84_markers.csv".
- The desired marker set should be activated by checking its box in the Marker Sets component. Here is an example using the data from the Hierarchical Clustering tutorial:
Retrieving selected annotations
- In the Marker Annotations component, select Retrieve Annotations. A portion of the returned results are shown below:
Displaying annotations and pathway diagrams
- The links under the heading Gene can be clicked to display information from the CGAP database at the NCI:
The Pathway links can be clicked to display BioCarta pathway diagrams provided through the NCI's caCORE/caBIO resource. The graphical components are themselves clickable to provide further information.
- Click a pathway link.
- Go to the caBIO Pathway viewer component. Below is displayed the pathway for the gene viewed above.
New Content
Marker Annotations The Marker Annotations component enables the retrieval of biological annotation information for a collection of genes. For every gene, the following data can be retrieved:
- A set of pathways containing the gene.
- A set of gene-disease and gene-compound associations derived from the literature articles.
All annotations are retrieved from remote servers maintained by the National Cancer Institute (NCI). The data in those server come from the following sources:
- Pathways: NCI's Pathway Interaction Database (PID). PID pathways come from 3 sources: BioCarta, Reactome and "NCI-Nature Curated". Information about the PID and each of the contributing sources is available at: http://pid.nci.nih.gov/userguide/database_content.shtml. These pathways are stored in servers used by the Cancer Gene Anatomy Project (CGAP, http://cgap.nci.nih.gov/).
- Gene-disease/compound associations: the Cancer Gene Index (CGI) data base. The reported assocations are extracted from article abstracts using a combination of automatic text mining, semi-automatic verification, and manual curation. Project details are available at: http://ncicb.nci.nih.gov/NCICB/projects/cgdcp.
Submit Query The Marker Annotations module will retrieve information for all markers that belong to activated marker sets, or, more precisely, for the genes corresponding to those markers:
E.g., in the example shown above, information will be retrieved about the genes AATF, CD40, STAT3. Checkboxes at the bottom of the component's user interface can be used to specify which data source(s) to query: CGAP, CGI or both. For CGAP, the associated drop-down can be used to designate the target organism for which annotations are retrieved: human (the default) or mouse. Clicking the "Retrieve Annotations" button initiates the communication with the NCI servers:
Pathway and Gene Annotations
The "Annotations" tab presents a summary listing of the annotations retrieved from CGAP:
The listing contains at least one row for each gene which annotation infromation is available for. If a gene is associated with more than one pathways, then one row for every pathway is listed (e.g., as is the case above for CD40 and STAT3). Every row displays the marker (i.e., probeset) id, the corresponding gene name and the name of the associated pathway. Clicking on a pathway brings up a popup menu offering a number options:
- View Diagram: available only for BioCarta pathways. Such pathways are accompanied by images offering a graphical/artistic rendition of the pathway. Selecting the "View Diagram" option will display this image wihtin the "Pathway" tab.
- Add pathway genes to set: extracts the pathway genes for which there are associated probes in the microrarray set currently selected by the user and places all such probes in a new marker set within the "Markers" component (by default, the marker set is named after the pathway).
- Export genes to CSV: creates a new text file containing a listing of all pathway genes. The file format (csv = comma separated values) is compatible with Microsoft Excel.
A BioCarta pathway image is displayed above after selecting the "View Dagram" option from the "Annotations" tab. The drop-down box, on the top left corner above the diagram, shows the name of the currently displayed diagram. The component keeps a history of all BioCarta diagrams selected by the user; using the drop-down it is possible to switch among the correponding pathway images. The "Clear Diagram" button clears the currently displayed diagram. The "Clear History" button both clears the currently displayed diagram and removes all pathway history information from the pathway name drop-down box.
In the "Annotations" tab, it is also possible to click on a gene name and explore functional annotation information from a number of sources (Entrez, CGAP, GeneCards):
Cancer Gene Index
For many genes, there are hundreds of records in the CGI database. Retrieving all those records at once can be a very time consuming operation, especially if the query invlolves many genes. To avoid very long waits, the retrieval of the data occurs in 2 stages. In the first stage, at most 10 records for each association type (gene-disease/gene-compound) are being fetched (for each query gene). Data retrieved as displayed in the CancerGeneIndex tab:
The user interface is divided in two regions, sharing the same overall structure and functionality: the table on the left displays gene-disease associations, while the table on the right is used for reporting gene-compound associations. To avoid redundancy, the paragraphs that follow describe only the gene-disease table. The exact same description applies in the case of the gene-compound table.
Each table row represent an association between a query gene and a disease. The first column contains the name of the probeset associated with the query gene. For any given gene, if its corresponding probeset name appears in bold-face, this means that there are more gene-disease records on the CGI server that have yet to be fetched (beyond the 10 records acquired in the initial retrieval stage). The disease name within a row is followed by a number in parentheses. This number indicates how many records (among those fetched) support the reported association. E.g, in the image above, the first row indicates that there are 6 distinct records linking STAT3 to melanoma. A detailed listing of these 6 records is available by "expanding" the row. This can be achieved by right-clicking on the row and selecting "expand" from the ensuing popup menu:
Each detailed record contains 3 additional pieces of information:
- Role: a curator-assigned description of the kind of association being reported. The values in this column come from a controlled vocabulary (developed to support the CGI database creation effort).
- Sentence: the actual article abstract sentence used to derive the reported gene-disease association. The full sentence is displayed in the text area at the bottom portion of the interface (it is also availalbe as a tooltip text, by mousing over the "Sentence" column).
- Pubmed: the Pubmed ID of the source article. Clicking on the Pubmed link brings up in the web browser the corresponding Pubmed abstract page (in this example, the sentence used for deriving the gene-disease association is actually the paper title):
It is also possible to link out to the NCI thesaurus in order to see the definition of the disease being associated with a gene; this is achieved by right-clicking on the table row for the gene-disease association and selecting the "Link to NCI_Thesaurus" option from the popup:
It should also be noted that the user interface allows ordering and filtering of the data (the latter can be very useful if there are many records being displayed):
- Ordering: the table rows can be sorted alphabetically by the contents of any column, by clicking on a column heading.
- Filtering: the drop down boxes that appear above the columns "Marker", "Gene", "Disease", and "Role" contain one value for each distinct entry within those columns. They can be used to select only records that contain the designated values. Of note is the drop-down associated with the "Disease" column:
The parantheses next to a disease name indicate how many distinct genes (among those included in the user query) are associated with this particular disease.
It should be noted that these numbers are calculated using only fetched records. As mentioned above, the first stage of the infromation retrieval will fetch at most 10 records per gene. The remaining records asscociated with a gene can be retrieved from the CGI server by right-clicking on a table row corresponding to the gene and by selecting the popup menu option "retrieve" all:
- IMAGE MISSING HERE****
After all gene-disease association records for a given gene have been fetched, the bold-face type of its associated probeset name is removed.
Finally,by clicking on the "Export" button at the bottom of the user interfce, the contents of the gene-disease and the gene-compound tables displayed within the CancerGeneIndex tab can be exported as comma separated values text files for further analysis or/and visualization by spreadsheet software.
Extra1
Extra2
Extra3