Difference between revisions of "Promoter Analysis"
(→The Results tab) |
(→Results) |
||
| Line 102: | Line 102: | ||
* Pseudocount - | * Pseudocount - | ||
| − | ==Results== | + | ===Results=== |
| − | ===Total hits and Seqeunces with hits=== | + | ====Total hits and Seqeunces with hits==== |
* Expected - | * Expected - | ||
* Actual - | * Actual - | ||
| Line 109: | Line 109: | ||
* % with hits | * % with hits | ||
| − | ===5' hits and 3'hits=== | + | ====5' hits and 3'hits==== |
* Expected - | * Expected - | ||
* Actual - | * Actual - | ||
Revision as of 14:07, 13 October 2009
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
Overview
The Promoter component scans one or more sequence profiles against nucleotide sequences that the user has loaded into geWorbench. Motifs from the JASPAR database of transcription factor binding sites are included with the component. Additional motifs can be added by the user.
The Promoter component will also display the results of hits found in the Pattern Discovery component.
JASPAR CORE database
The motif datafile currently included with geWorkbench is version 3.0 of the JASPAR CORE database (http://jaspar.genereg.net/) . It contains 138 curated, non-redundant profiles. These "profiles are derived from published collections of experimentally defined transcription factor binding sites for multi-cellular eukaryotes. The database represents a curated collection of target sequences" (JASPAR Documentation).
The datafile used "MATRIX_DATA.txt" found at http://jaspar.genereg.net/html/DOWNLOAD/mySQL/JASPAR_CORE_2008/. This file is stored within the Promoter component of geWorkbench.
The profiles are represented by counts of how many times each of the four nucleotide bases occurs at a particular position in the aligned sequences.
Working with the Promoter graphical interface
Layout
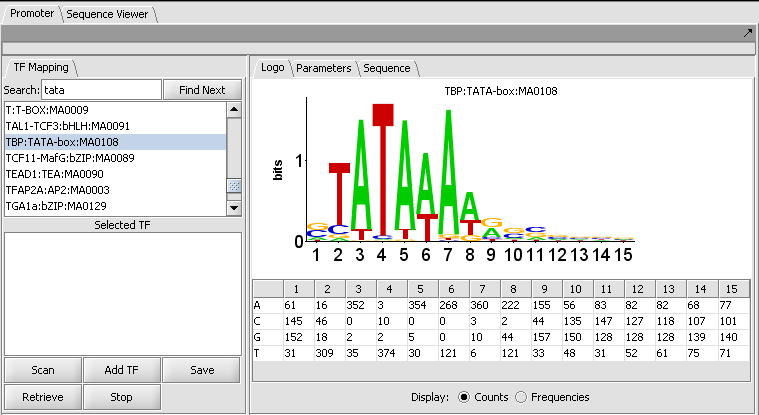
The figure shows the display of profile "TBP:TATA-box:MA0108" from the list of those included in JASPAR.
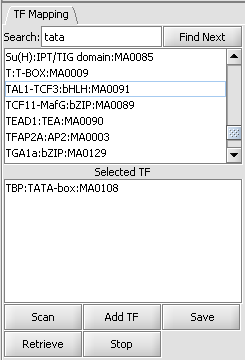
TF Mapping
List of available transcription factors
This list contains all loaded transcription factor profiles, including those from JASPAR and any loaded by the user. A profile can be transferred to the "Selected TF" list just below by double-clicking on its entry. Multiple profiles can be moved to the selected list.
- Search - search the list items for the input characters strings.
- Find Next - continue scanning the list for the next occurrence of the search string.
Selected TF
This list contains the profiles that will be used in the next scan. Entries on the Selected TF list can be moved back up to the "available TFs" list by again double-clicking on their entry.
Here the TATA-box entry has been moved to the selected list.
Controls
- Scan - start the scan of the current nucleotide sequence with the selected profiles.
- Add TF - load a new profile from a file into the list of available profiles. This is not a permanent addition; it remains loaded only for the current invocation of geWorkbench. See the "Profile File Format" entry below for details.
- Save - Saves a list of hits by a profile to a nucleotide sequence, including the sequence identifier and the start and stop points of the match along the sequence, as shown here:
gi|65508003
ATHB5:HOMEO-ZIP:MA0110 2104 2113
ATHB5:HOMEO-ZIP:MA0110 2115 2106
ATHB5:HOMEO-ZIP:MA0110 2882 2891
ATHB5:HOMEO-ZIP:MA0110 2893 2884
- Retrieve - Not implemented.
- Stop - Stop the current scan.
Profile file format
A profile in the form of a count matrix can be loaded from an external file. The profile should consist of a tab delimited series of counts, one for each position in the profile. It should could four lines, in the order A, C, G, T. There are no header lines or row labels, just the numeric matrix. For example, here is a profile showing the first six columns:
0 12 0 0 1 0
49 0 20 23 3 45
0 37 29 2 45 4
0 0 0 25 0 0
Because the normalization step uses the count total (sequences aligned to generate the profile), loading a frequency matrix is not currently supported.
The LOGO tab
LOGO display
The LOGO display implements the method of Schneider and Stephens (1990) to display the information at each position in a motif. Briefly, the total height of the column of letters at a position shows the information available, on a scale of 0 to 2 bits (the information needed to represent the 4 possible nucleotide bases at each position). The relative heights of each letter in a column show their individual contribution to the information at that position.
The LOGO display in geWorkbench implements the "small sample correction" described by Schneider, the magnitude of which depends on the number of sequences aligned to generate the profile. The correction is subtracted from the calculated information content at each position, with a minimum value (floor) of zero being displayed.
Table display
A table is used to show the numeric data from which the LOGO diagram is generated. The table depicts each position in the profile as a column, and has a row for each of the four nucleotide bases A, C, G and T. The user can choose to display the data either as the original counts or as frequencies.
- Display: Counts or Frequencies
The Parameters tab
Parameters
- PValue / 1K -
- Use Thr. - Use threshold -
- 13K Set -
- Iterations -
- Pseudocount -
Results
Total hits and Seqeunces with hits
- Expected -
- Actual -
- Enrich. p-value -
- % with hits
5' hits and 3'hits
- Expected -
- Actual -
The Sequence tab
Scan Implementation
Normalization and the Pseudocount
The count matrices are normalized to frequencies using an algorithm which includes a "pseudocount" (see Nishida 2009). The pseudocount is a way to compensate for the effects of small sample sizes in the original observations used to generate the profiles. Nishida et al. studied how to determine an appropriate value for the pseudocount. They found that the optimal values were independent of the sample size and were correlated with the entropy of the original matrices. They say that this implies that the less-conserved the binding site, the larger a value should be used for the pseudocount. They find that 0.8 is a good value "for practical uses". They do not recommend use of the square root of the total count.
geWorkbench allows a pseudocount factor to be directly entered, or it can be selected to be the square root of the total count of sequences used to generate the profile. Prior to geWorkbench 1.8.0, setting the pseudocount to the square root of the total counts was directly coded and not changeable. The current default is to set the pseudocount equal to 1.0.
The normalization forumula used in calculating frequencies is then, where b is the pseudocount, and counts(i, j) is the observed count in a particular entry in the matrix,
freq(i, j) = (counts(i, j) + b/4) / (totalCounts + b).
The resulting frequency matrix is used in the subsequent scan.
Because the pseudocount is a settable parameter, the frequency matrix is recalculated for each scan from the original counts.
Running and viewing a scan
- Hit Scan.
- Check the box "Show TFs".
- Hits of this motif against the sequences are displayed in the sequence window.
References
- Nishida K, Frith MC, Nakai K. (2009) Pseudocounts for transcription factor binding sites. Nucleic Acids Res. Feb;37(3):939-44. link to paper
- Sandelin A, Alkema W, Engstrom P, Wasserman WW, Lenhard B. (2004) JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. Jan 1;32(Database issue):D91-4 (link to paper).
- Schneider TD, Stephens RM. (1990) Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. Oct 25;18(20):6097-100. (link to paper)
- Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW, van Roy F, Lenhard B. (2006) A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res. Jan 1;34(Database issue):D95-7.