Difference between revisions of "CaArray"
(→Searching and viewing available experiments) |
(→The Remote Open File dialog) |
||
| Line 2: | Line 2: | ||
| − | =The Remote Open File dialog= | + | =The Remote Open File dialog (caArray)= |
| − | geWorkbench can retrieve data from | + | geWorkbench can retrieve gene expression data from remote instances of the NCICB's caArray database. These may be copies maintained by the NCI itself, or copies maintained locally at your or another institution. You can maintain settings for any number of different caArray installations. |
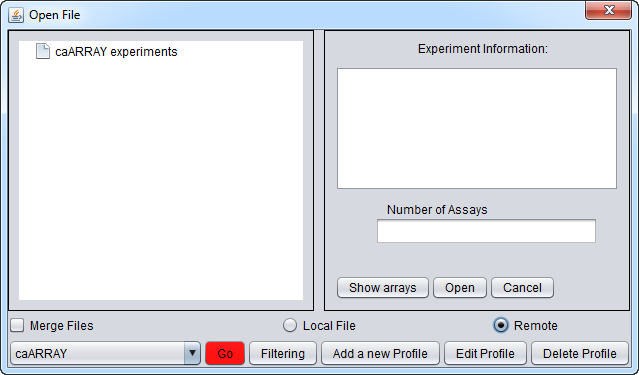
Right-click on '''Project''' which will bring up the '''Open File''' dialog. Click the '''Remote''' radio button. The '''Open File''' dialog window will be expanded to include remote sources. | Right-click on '''Project''' which will bring up the '''Open File''' dialog. Click the '''Remote''' radio button. The '''Open File''' dialog window will be expanded to include remote sources. | ||
| Line 13: | Line 13: | ||
They buttons at the bottom of the remote file dialog are: | They buttons at the bottom of the remote file dialog are: | ||
| − | '''Source''' | + | '''caArray (Source)''' menu - Shows a list of caArray instances that have been configured. Entries for the "Production" and "Stage" instances of caArray maintained by NCI are preconfigured. |
'''Go''' button - downloads a list of all available experiments from the remote source. | '''Go''' button - downloads a list of all available experiments from the remote source. | ||
| − | '''Filtering''' - | + | '''Filtering''' - The list of available experiments can be "filtered" to shown only those matching particular criteria. The available options are: |
| + | * Categories: Experiment | ||
| + | * Field Selection: Array Provider, Organism, Principal Investigator | ||
| + | * Values: | ||
| + | ** Array Provider: Affymetrix, Agilent, GenePix, Illumina, ImaGene, Niblegen, ScanArray, UCSF Spot. | ||
| + | ** Organism: many entries, including human, mouse, fly etc... | ||
| + | **Principal Investigator: PIs of listed public experiments in the particular instance of caArray. | ||
| − | '''Add A New | + | '''Add A New Profile''' button - Opens the Data Source Definition Page used to add a new instance of caArray. |
| − | |||
| − | |||
| + | '''Edit Profile''' button - Edit the currently selected profile. | ||
| + | '''Delete Profile''' - Remove the currently selected profile. | ||
=Loading data from an instance of caArray= | =Loading data from an instance of caArray= | ||
Revision as of 12:53, 15 June 2010
Contents
The Remote Open File dialog (caArray)
geWorkbench can retrieve gene expression data from remote instances of the NCICB's caArray database. These may be copies maintained by the NCI itself, or copies maintained locally at your or another institution. You can maintain settings for any number of different caArray installations.
Right-click on Project which will bring up the Open File dialog. Click the Remote radio button. The Open File dialog window will be expanded to include remote sources.
They buttons at the bottom of the remote file dialog are:
caArray (Source) menu - Shows a list of caArray instances that have been configured. Entries for the "Production" and "Stage" instances of caArray maintained by NCI are preconfigured.
Go button - downloads a list of all available experiments from the remote source.
Filtering - The list of available experiments can be "filtered" to shown only those matching particular criteria. The available options are:
- Categories: Experiment
- Field Selection: Array Provider, Organism, Principal Investigator
- Values:
- Array Provider: Affymetrix, Agilent, GenePix, Illumina, ImaGene, Niblegen, ScanArray, UCSF Spot.
- Organism: many entries, including human, mouse, fly etc...
- Principal Investigator: PIs of listed public experiments in the particular instance of caArray.
Add A New Profile button - Opens the Data Source Definition Page used to add a new instance of caArray.
Edit Profile button - Edit the currently selected profile.
Delete Profile - Remove the currently selected profile.
Loading data from an instance of caArray
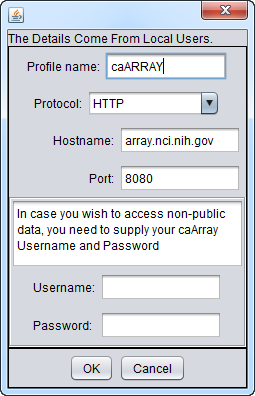
Setting up the connection
You can Add a New Resource or Edit existing connection settings to set up a connection to an instance of caArray. The configuration for connecting to the production instance of caArray at the NCI is shown here:
Searching and viewing available experiments
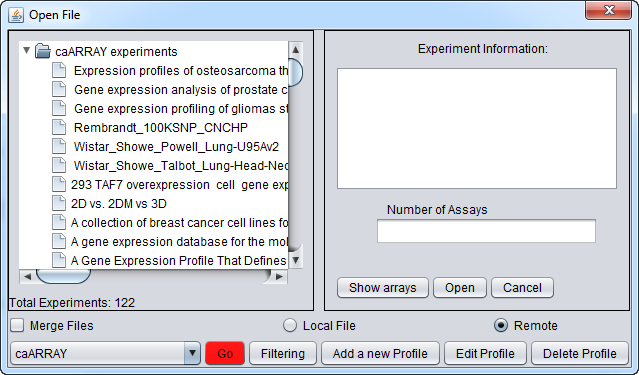
If you click on the red Go button next to the caArray data source at the bottom of the dialog, all available caArray experiments at that location will be displayed.
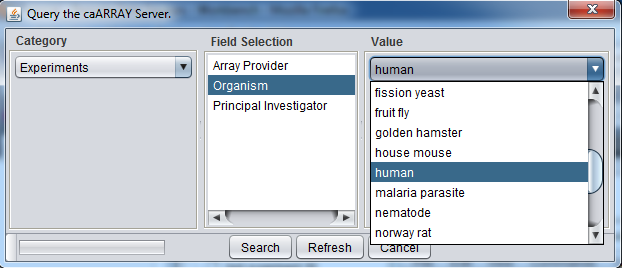
Instead, you can select only particular kinds of experiments by pushing the Filter button. Here we show experiments of type "Human" being selected.
And here are the resulting entries in the database:
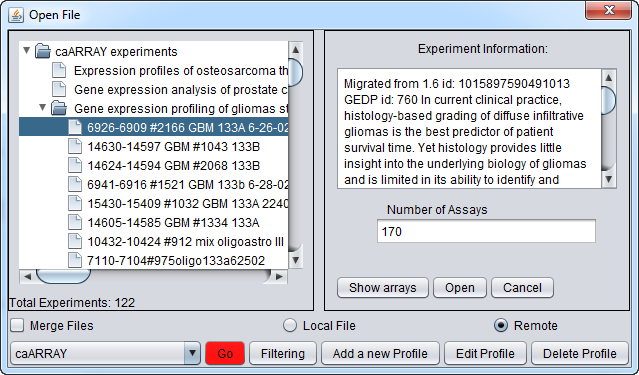
Select an experiment and push the Show Arrays button to see the individual array datasets available for download for this experiment.
Downloading select array datasets
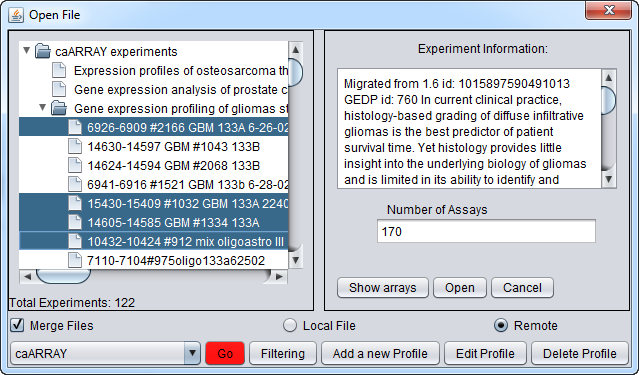
Now we will select four of the arrays of type HG-U133A and push the Open button to begin the download. Dont' forget to click the Merge button first if desired to merge the data into a single dataset.
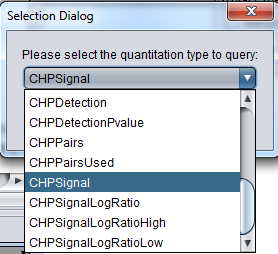
You will be prompted to select the quantitation type from those available for the experiment. Here we select CHP Signal:
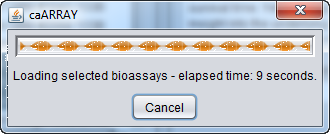
A progress bar will track the download process:
The resulting data set will appear in the Project Folders component: