Difference between revisions of "Cellular Networks KnowledgeBase"
(→Overview) |
(→Marker) |
||
| Line 44: | Line 44: | ||
===Marker=== | ===Marker=== | ||
| − | The marker name (comes from the microarray set node selected in the Project Folders area). | + | The marker (probeset) name (comes from the microarray set node selected in the Project Folders area). |
| + | |||
===Gene=== | ===Gene=== | ||
The gene name corresponding to the marker, if loaded from an annotation file. | The gene name corresponding to the marker, if loaded from an annotation file. | ||
Revision as of 14:24, 28 January 2011
|
Home | Quick Start | Basics | Menu Bar | Preferences | Component Configuration Manager | Workspace | Information Panel | Local Data Files | File Formats | caArray | Array Sets | Marker Sets | Microarray Dataset Viewers | Filtering | Normalization | Tutorial Data | geWorkbench-web Tutorials |
Analysis Framework | ANOVA | ARACNe | BLAST | Cellular Networks KnowledgeBase | CeRNA/Hermes Query | Classification (KNN, WV) | Color Mosaic | Consensus Clustering | Cytoscape | Cupid | DeMAND | Expression Value Distribution | Fold-Change | Gene Ontology Term Analysis | Gene Ontology Viewer | GenomeSpace | genSpace | Grid Services | GSEA | Hierarchical Clustering | IDEA | Jmol | K-Means Clustering | LINCS Query | Marker Annotations | MarkUs | Master Regulator Analysis | (MRA-FET Method) | (MRA-MARINa Method) | MatrixREDUCE | MINDy | Pattern Discovery | PCA | Promoter Analysis | Pudge | SAM | Sequence Retriever | SkyBase | SkyLine | SOM | SVM | T-Test | Viper Analysis | Volcano Plot |
Contents
- 1 Overview
- 2 Working with the CNKB graphical interface
- 3 Example - Creating an interaction network and viewing it in Cytoscape
- 4 Appendix - Data Sources
- 5 References
Overview
This material describes the graphical interface and functionality of the CNKB component released with geWorkbench 2.1.0. Significant enhancements have been made with each recent release.
General
The Cellular Network Knowledge Base (CNKB) is a repository of molecular interactions, including ones both computationally and experimentally derived. Sources for interactions include both publicly available databases such as BioGRID and HPRD, as well as reverse-engineered cellular regulatory interactomes developed in the lab of Dr. Andrea Califano at Columbia University.
Each pairwise interaction may have an associated confidence indicator (a value between 0 and 1) or another dataset-specific metric reflecting the strength of the underlying data, whether experimental or computational. Details on the methodology used to construct the CNKB are available in Mani et al. 2008.
Gene interaction information from the CNKB can be used, for example, in order to assess the plausibility of a hypothesis of concerted molecular action represented by a gene set that has been discovered using computational approaches (e.g., by running a clustering analysis on a microarray set to identify tandems of co-expressed genes). If the genes in such a set are reported in the CNKB to have several direct interactions (or several common targets) then this may be evidence that the gene set indeed reflects at some level a real biological process.
The CNKB component allows the user to select a group of markers of interest, specify the interaction types (e.g. Protein-Protein, Protein-DNA etc.), and choose a particular interaction data source. After a query of the Knowledge Base, results are displayed both in the CNKB component and as interaction graphs in the Cytoscape component.
The CNKB graphical interface
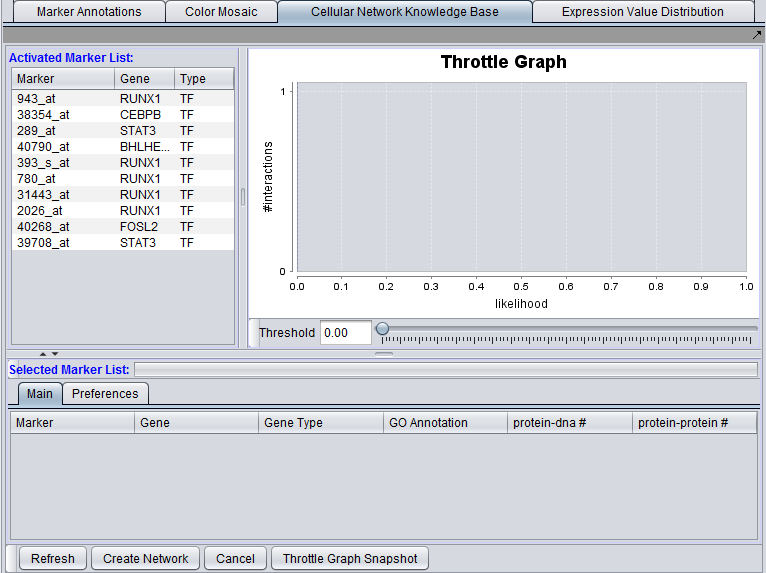
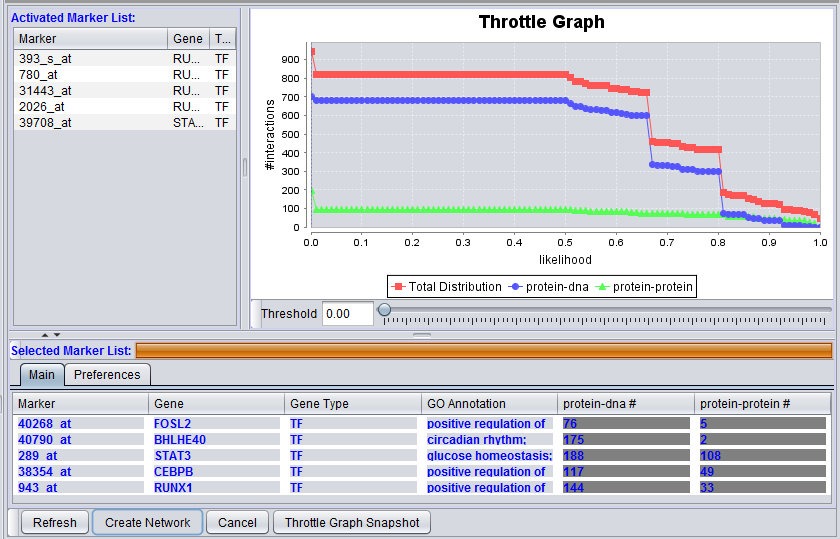
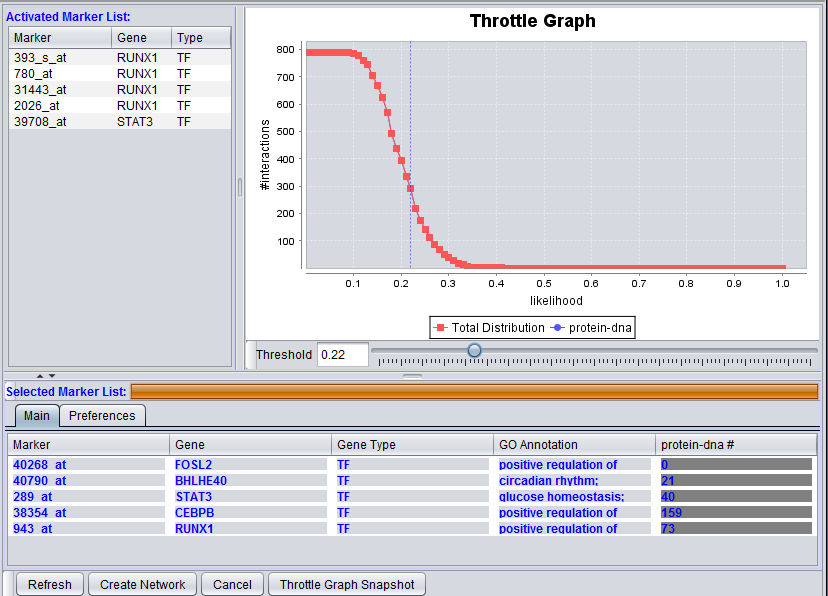
The graphical user interface (GUI) of the CNKB component has three areas of distinct functionality: The "Activated Marker List", the "Selected Marker List", and the "Throttle Graph". The data source, version, and interaction types are specified on a separate "Preferences" tab.
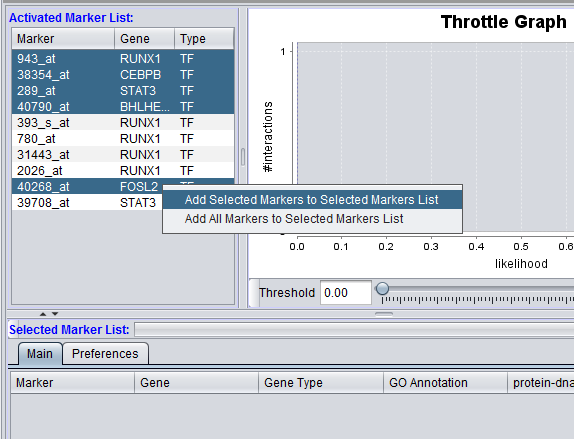
The "Activated Marker List" displays markers activated in the Markers component. From this list, the user can select markers to use in querying the CNKB database. This can be done either by double-clicking on desired markers, or through use of a right-click menu (see below); those selected will be added to the "Selected Marker List" just below.
The "Selected Marker List" displays which markers will be used in the query, and also displays the query results.
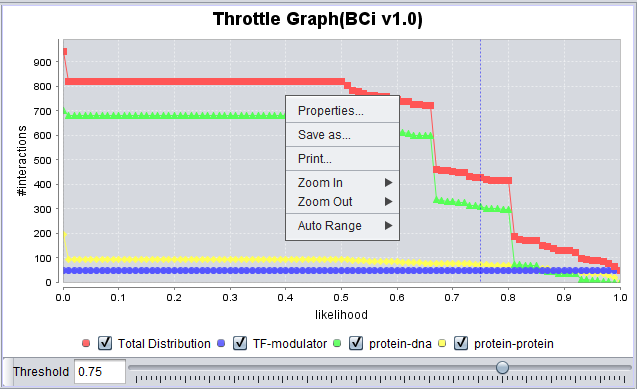
Interactions which will be used to build a network graph can be adjusted using the "Throttle Graph". It is used to set a minimum confidence threshold for inclusion in the network.
A query against the CNKB database is initiated by clicking on the "Refresh" button. All pairwise interactions in the chosen data source of the desired types(s) that involve any marker in the "Selected Marker List" are retrieved.
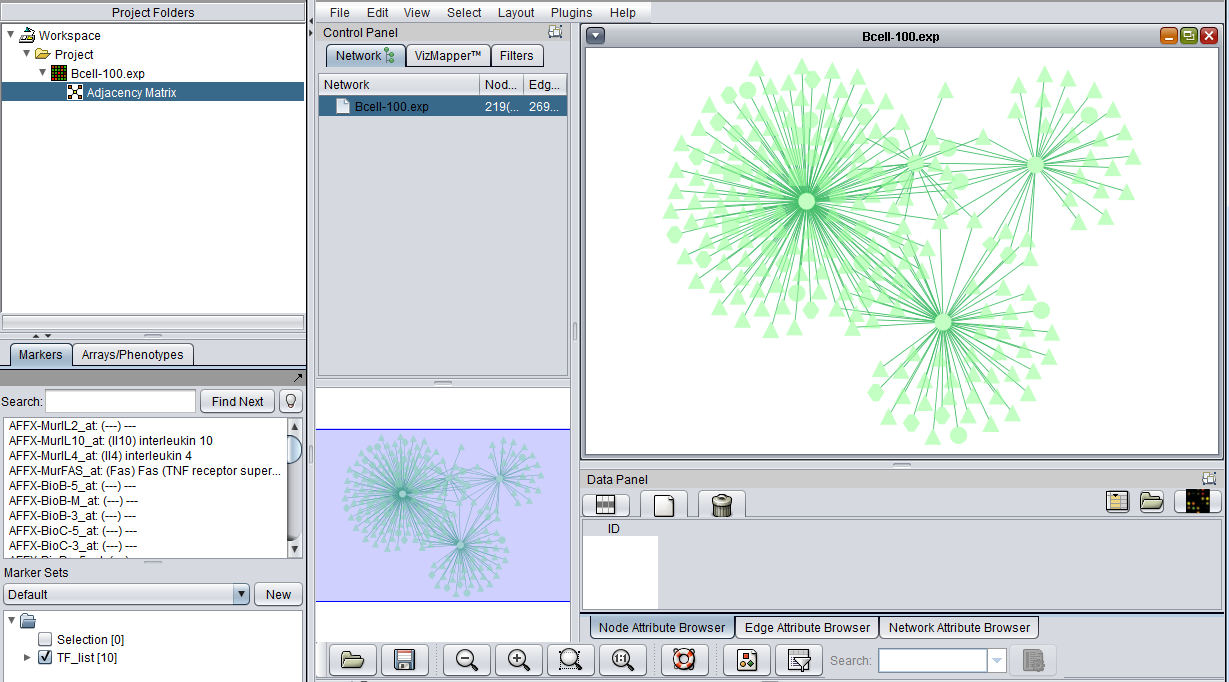
An interaction network can be constructed by pushing the "Create Network" button. The network will be placed as an adjacency matrix in the Project Folders component and will be automatically displayed in the Cytoscape component.
Working with the CNKB graphical interface
The figures in this section correspond to the example in the final section of this tutorial.
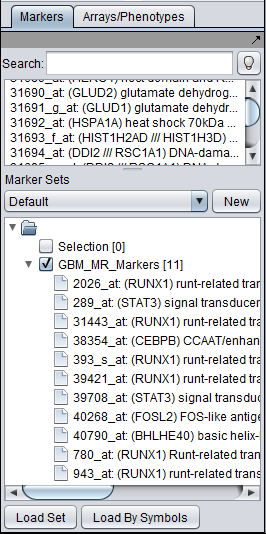
Activated Marker List
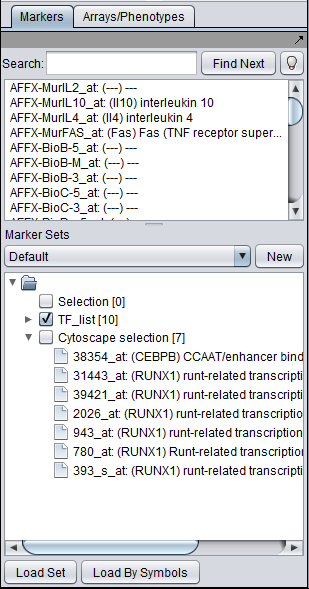
The "Activated Marker List" contains the markers that belong to activated marker sets from the Markers component. It contains 3 columns:
Marker
The marker (probeset) name (comes from the microarray set node selected in the Project Folders area).
Gene
The gene name corresponding to the marker, if loaded from an annotation file.
Type
A gene type designation, derived from the gene's GO annotation:
- TF - Transcription Factor,
- K - Kinase,
- P - Phosphatase, and
- (no entry) - type is unknown.
Individual markers in the Activated Marker List can be moved to the Selected Markers List by double-clicking on them.
Alternatively, a right-click menu (shown above) allow a group of highlighted markers, or all markers, to be moved to the "Selected Markers List".
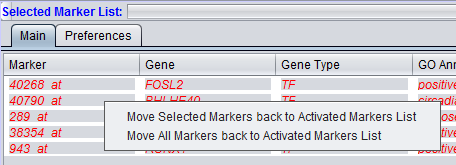
Selected Marker List
Main
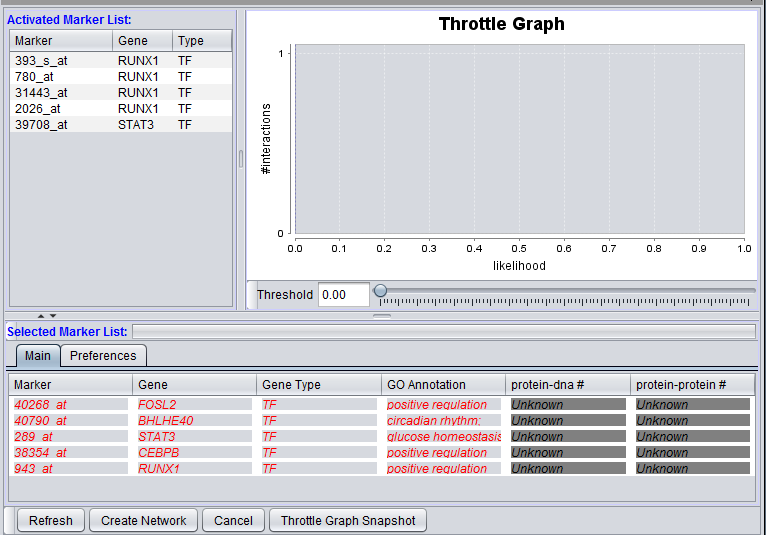
Markers which have been moved from the Activated Markers List to the Selected Markers List can be used to query the CNKB database. Until a query is run, the list items are shown in red and in italics.
After a query has been run against the CNKB database, the marker entries are shown in regular font, blue letters.
Items that have been added to the Selected Markers list can be removed and sent back to the Activated Markers list by double clicking on their entries or through a right-click menu. The right-click menu (shown below) gives the choice of moving only highlighted, or all markers back to the Activated Markers list.
The Selected Marker List table contains the following columns:
Marker
The marker name (same as in the "Activated Marker List"). Appears italicized to indicate that interaction information has not yet been retrieved. Bold face font indicates that interaction information has been retrieved (and is displayed).
Gene
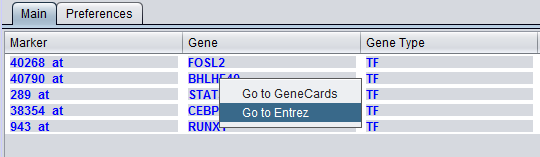
The gene name corresponding to the marker, if loaded from an annotation file (same as in the "Activated Marker List").
Right-clicking on a gene name will provide link-outs to Gene Cards and Entrez Gene.
Gene Type
A gene type designation, derived from the gene's GO annotation The list of type codes is the same as that under Type under "Activated Marker List".
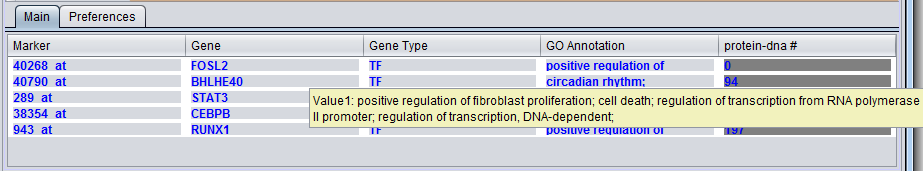
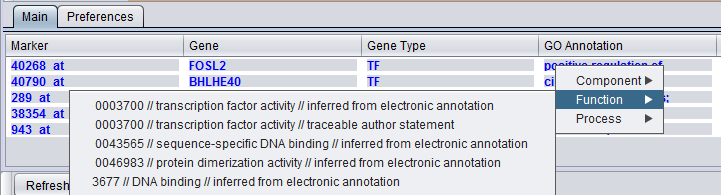
GO Annotation
The GO annotation of the gene. Right clicking on the column brings up the the gene GO classification of the gene across the 3 top-level GO categories: Component, Function and Process.
Hovering the mouse cursor over an entry in the GO Description column will display a short summary of the Gene Ontology terms associated with that entry.
More extensive GO annotations can be viewed for desired genes in the Selected Marker list by right-clicking on its entry. A pop-up menu will offer a choice of the three categories of GO annotation: Component, Function and Process. Expanding one of these terms will show the available annotations for that gene.
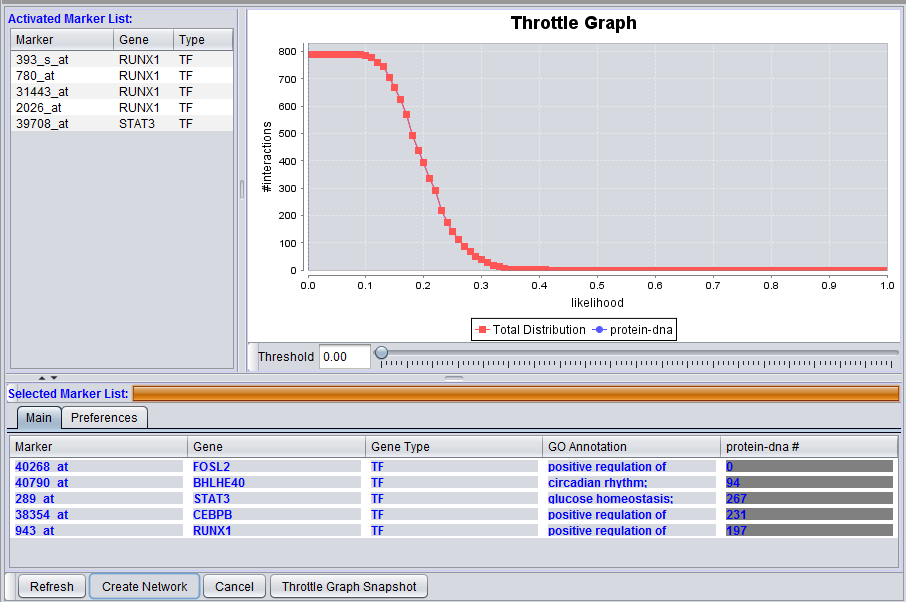
Interaction Query columns
A column will display interaction results for each data source selected in the Preferences tab. It will show, for each marker, the number of interactions that meet the threshold currently set in the throttle graph slider control.
The illustrations above depict a Protein-DNA interaction column.
Preferences
Interactions Database
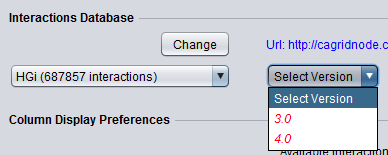
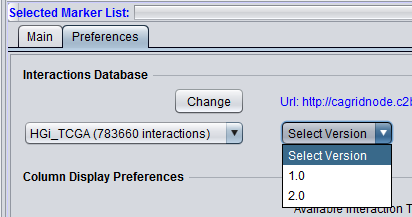
Here the desired interaction data source and version can be chosen.
Change - This button allows the address of the CNKB servlet to be changed. This should not normally be necessary.
Database pulldown menu - This menu contains all data sources available via the CNKB component. Only databases that have interactions (not empty) are displayed. The number of interactions is also displayed.
Select Version - Each data source has a version associated with it. Versions may be updates or may contain different types of interactions from a particular system. The exact contents of each data source are available on the CNKB data sources page.
Some interactomes may contain datasets not yet made public. These will be shown in red and require a password to access them.
Here we will use the public interactome of HGi_TCGA, version 1.
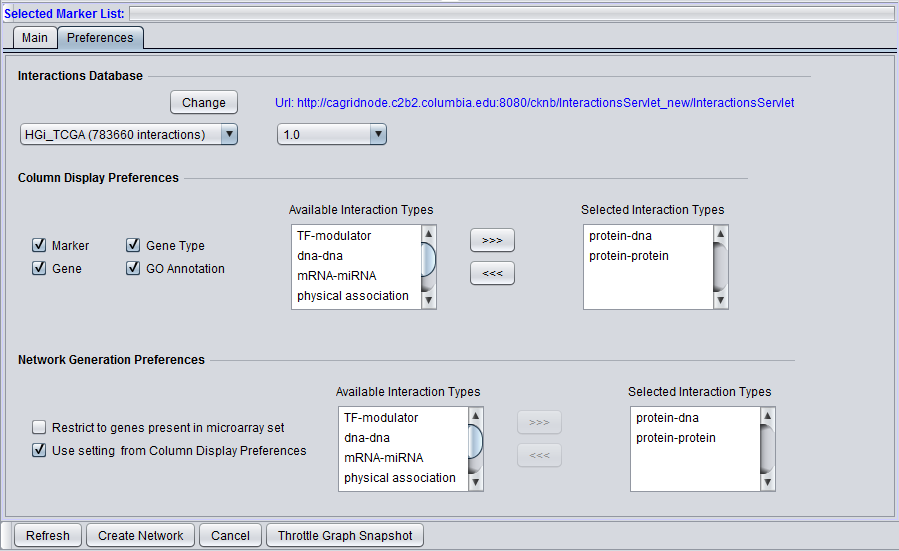
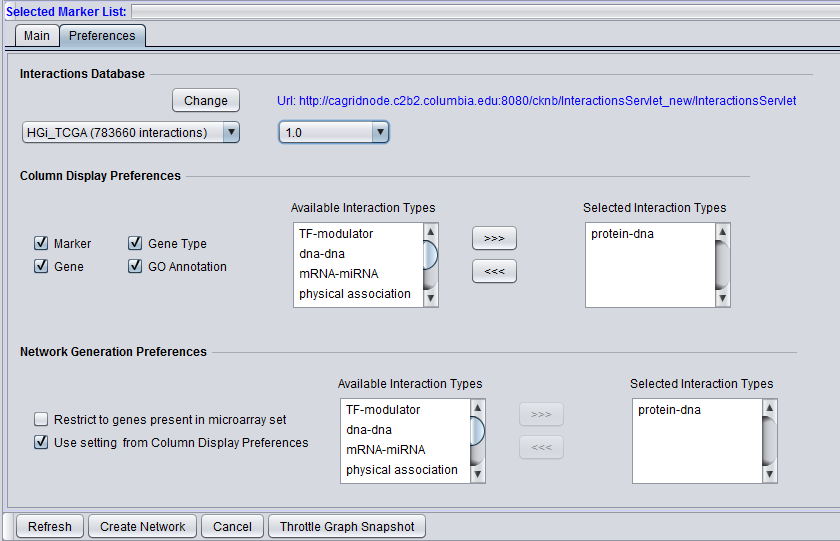
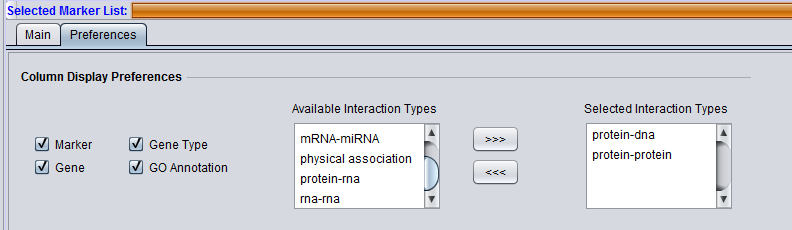
Column Display Preferences
These selections control which data columns will be displayed in the main "Selected Markers" pane.
- Marker
- Gene
- Gene Type
- GO Annotation
- Available Interaction Types - Contains a list of all interaction types used in the CNKB component that have not been already moved to the "Selected Interaction Types" list to the right. Note that this list is not specific to the particular data source chosen.
- Selected Interaction Types - A column in the Main tab "Selected Marker List" will appear for each interaction type appearing in this list.
List entries can be moved between the "Available" and "Selected" lists by either double-clicking directly on an entry, or through use of the right and left triple arrows "<<<", ">>>" located between them.
Network Generation Preferences
This section controls how the interactions returned by the database query will be used in creating a network. It allows, if desired, separate control of query and display.
- Restrict to genes present in microarray set - queries to the CNKB database may return interaction partners that are not members of the microarray dataset from which the original query markers were chosen. Checking this box will cause such markers NOT to be used in generating a network graph.
- Use setting from column display preferences
- If checked, the same interaction types that have been selected in the "Column Display Preferences" control above will be used in generating the network graph.
- If unchecked, only those interaction types added to the "Selected Interaction Types" list will be used to construct a network graph.
Throttle Graph
This interactive graph allows users to "throttle" (for the genes in the Selected Markers table) which interactions to work with, using as a criterion the interactions’ confidence indicator. As the required threshold of likelihood of the interactions is increased, the sum of interactions meeting this criterion decreases.
- Right-click menu - Right-clicking on the Throttle Graph will bring up a menu which allows the graph to be customized.
- Properties -
- Save as -
- Print -
- Zoom in -
- Zoom out -
- Auto Range - return to an automatically calculated range e.g. after use of Zoom function.
The BCi V1.0 (Bcell interactome V1.0) has both Protein-DNA and Protein-Protein interaction types. If we select both of those types in the Preferences tab,
then two results columns appear in the results table, and both interaction types are shown as different color lines in the Throttle Graph.
Example - Creating an interaction network and viewing it in Cytoscape
Once the desired set of markers is present and its interaction data has been retrieved from the database, an adjacency matrix can be computed by clicking the "Create Network" button. The resulting matrix is placed in the Project Folders component under its parent microarray dataset. The adjacency matrix is visualized in the Cytoscape Viewer.
Please note that if the targets retrieved from the Knowledge Base include markers/genes not present in the active microarray dataset, then whether those markers will be used in creating the network graph in Cytoscape is determined by the setting of the "Restrict to genes present in microarray set" checkbox under "Network Generation Preferences" on the CNKB Preferences tab.
The Cytoscape Viewer maintains a list of networks which it has currently loaded. It allows individual loaded networks to be deleted. However, the network can be reloaded by clicking on its entry in the Project Folders component. Cytoscape controls are more fully described in the Cytoscape component tutorial.
Prerequisites
- This example uses the Bcell-100.exp dataset available in the data/public_data directory of geWorkbench, and further described on the Download page. Briefly, this dataset is composed of 100 Affymetrix HG-U95Av2 arrays on which various B-cell lines, both normal and cancerous, were analyzed. Thus it explores a potentially wide variety of expression phenotypes.
- Obtain the annotation file for the HG-U95Av2 array type from the Affymetrix NetAffx website (http://www.affymetrix.com/analysis/index.affx). The name will be similar to "HG_U95Av2.na30.annot.csv", where "na30" is the version number. Loading the annotation file associates gene names and Gene Ontology information with the Affymetrix probeset IDs (see the geWorkbench FAQ for details on obtaining these files).
Loading the example data
- Load the Bcell-100.exp dataset into geWorkbench as type "Affymetrix File Matrix". (See Local Data Files).
- When prompted, load the annotation file.
- Create and activate a set of markers in the Markers component. For this example, save the file GBM_MR_Markers to disk and then load it directly into the Markers component by pushing its "Load Set" button and browsing to the file located in the geWorkbench directory data/public_data.
When the set is activated by checking the box to the left of its name, the transcription factor markers will appear in the "Activate Markers" list in the CNKB component.
Setting up the query in the CNKB component
(The steps shown below are also depicted in the screenshots for the individual control descriptions above).
1. On the CNKB Main tab, right-clicking on the "Activated Markers" list will bring up a menu which allows one to move all activated markers or just the highlighted markers to the "Selected Markers List" for querying.
2. Select the desired data source and interaction types in the CNKB Preferences tab.
3. Now hit the "Refresh" button to perform the query against the Cellular Networks Knowledge Base database.
4. Adjust the Throttle Graph allows to set a minimum confidence requirement on interactions that will be used to create a network. In the example images above, a value of 0.22 was used.
Note how the number of interactions is decreased as the threshold is increased.
5. Hit the "Create Network" button.
6. The resulting adjacency matrix is displayed in the Cytoscape component.
Integration of Cytoscape and geWorkbench
The use of Cytoscape for network visualization is covered in detail in the tutorial Tutorial_-_Cytoscape_Network_Viewer. Here we show a few ways in which the Cytoscape component can be used to investigate the interaction results.
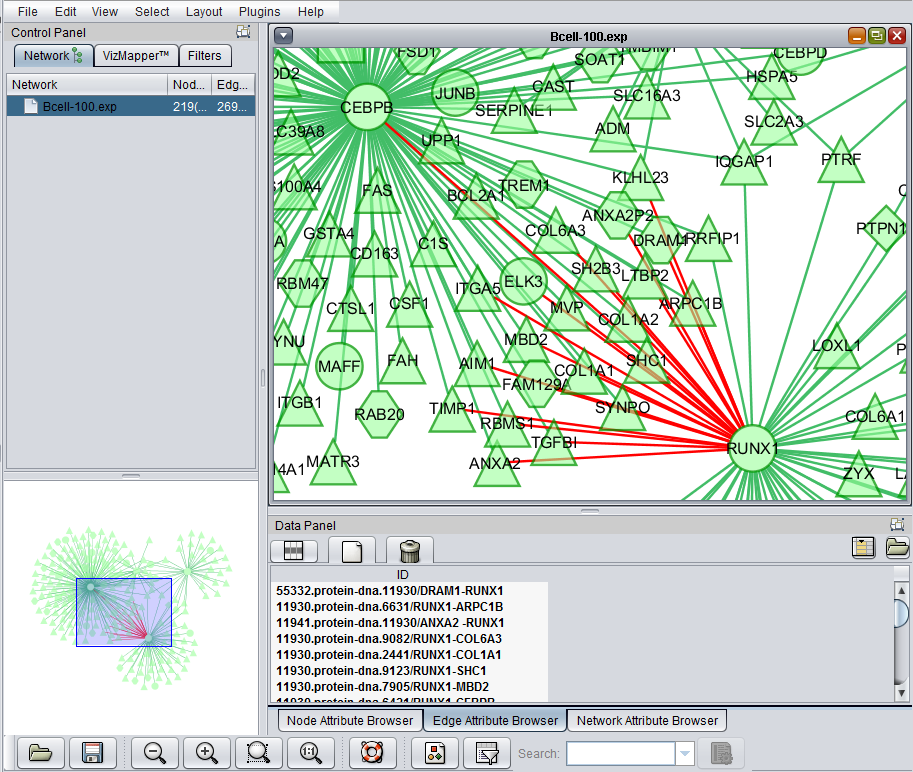
Selecting interactions (edges)
Using the mouse, a group of edges can be selected.
The list of selected edges is displayed in a list below the graph, in a tab titled "Edge Attribute Browser".
Selecting nodes
Multiple nodes/genes can be selected by holding down the Shift key while left-clicking on individual nodes.
The markers corresponding to the selected genes will be displayed directly in the Markers component in a new subset called "Cytoscape selection". Note that this set is dynamically updated - it displays markers corresponding to whatever nodes are currently highlighted in the network graph.
To make a permanent copy of a particular set of dynamically chosen markers in the "Cytoscape Selection" subset, right-click on it and select "Copy".
Options for selected nodes
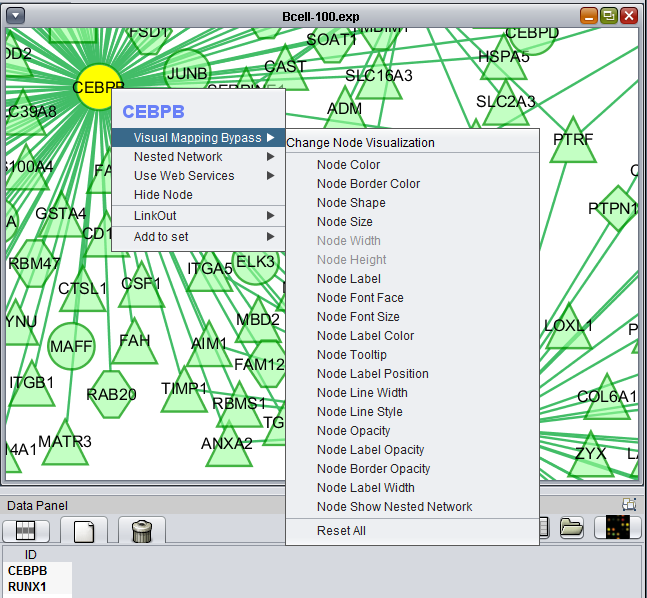
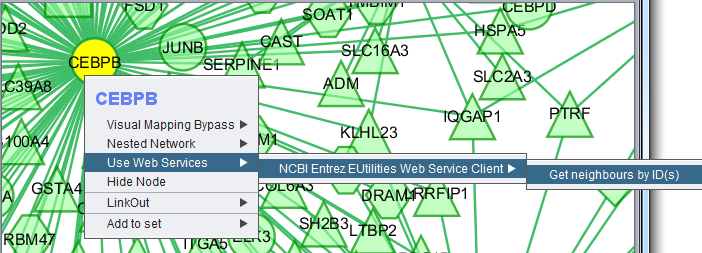
Right-clicking on a particular node in the network graph brings up a menu with three options:
- Visual Mapping Bypass
- LinkOut
- Add to set
Visual Mapping Bypass
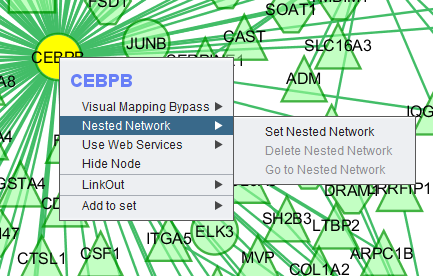
Nested Network
Use Web Services
Hide Node
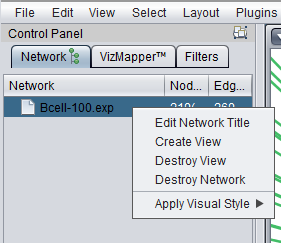
Selecting "Hide Node" will cause any selected nodes and all their edges to disappear.
So far, the only way we have found to rematerialize a hidden node is to go the Cytoscape Network tab, select the network, destroy its view and then create a new view.
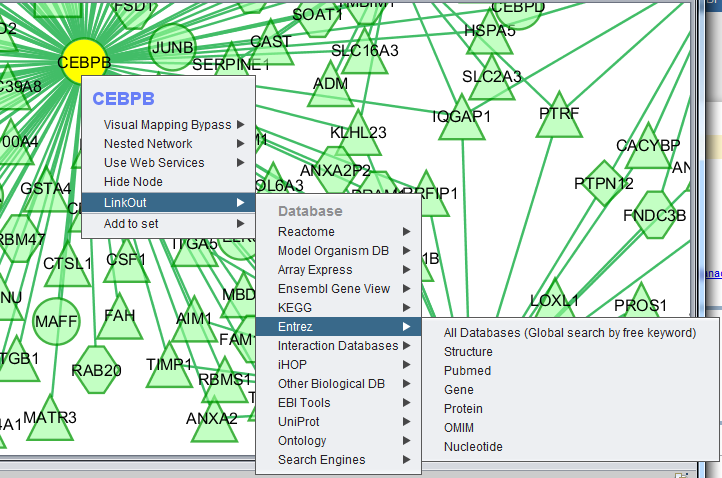
LinkOut
This menu option provides hyperlinks to a number of external sources of gene annotation.
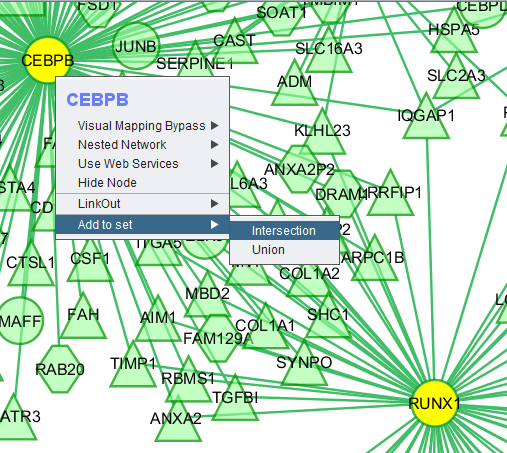
Add to set
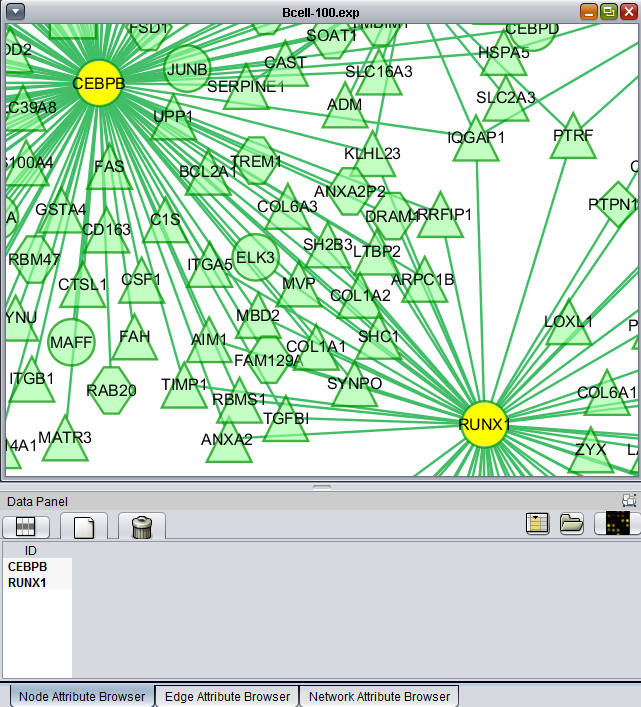
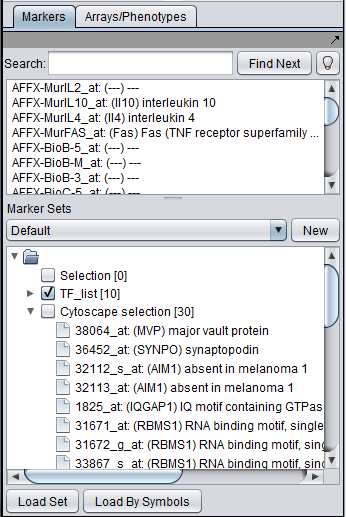
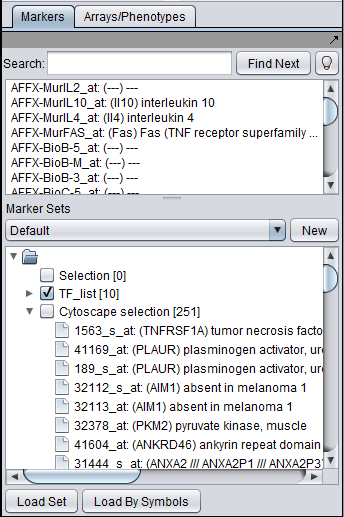
If one or more graph nodes have been selected (highlighted in yellow in figure below), the markers they directly interact with (via edges) can be copied to the default "Cytoscape selection" subset in the Markers component at lower left in the geWorkbench graphical interface.
Two options are available under "Add to Set". These are
- Intersection - find the set of markers that have interactions (edges) with ALL selected nodes. Such markers are placed into the Markers component, in the "Cytoscape selection" subset.
- Union - find the set of markers that have interactions (edges) with ANY of the selected nodes. Such markers are placed into the Markers component, in the "Cytoscape selection" subset.
This image shows the intersection set of markers for the two selected genes:
This image shows the union set of markers for the two selected genes.
Appendix - Data Sources
Please see the CNKB data page for a list of currently available data sources and types of interactions.
References
- Mani KM, Lefebvre C, Wang K, Lim WK, Basso K, Dalla-Favera R, Califano A., " A systems biology approach to prediction of oncogenes and molecular perturbation targets in B-cell lymphomas", Molecular Systems Biology 4:169, 2008. link to paper