Array Sets
Contents
Preparation
In this tutorial we will start with the same data files that were used in Tutorial - Projects and Data Files. Load the ten individual MAS5 data files as shown there.
Creating sets
When working with microarrays, geWorkBench uses the term marker to refer to a gene probe (in other cases, it can be individual items from other data sets, such as sequences).
Phenotype refers to any user-defined grouping of microarrays. These microarrays will typically share some common property. For example, two such phenotypes might be the diseased and normal states of a tissue from which samples have been taken.
Assigning arrays to sets
geWorkbench supports groupings of sets. We will place the phenotypes in the default group, however you can create new phenotype group by pushing the New button on Phenotype Sets at lower left.
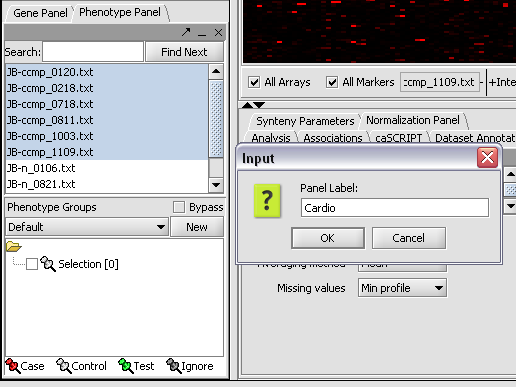
First, we will select and label arrays which contain samples from the congestive cardiomyopathy disease state:
1. In the Arrays/Phenotypes component, select the following arrays from samples from the congestive cardiomyopathy disease state:
JB-ccmp_0120.txt, JB-ccmp_0218.txt, JB-ccmp0718.txt, JB-ccmp0811.txt, JB-ccmp1003.txt, JB-ccmp_1109.tx
2. Right-click, select Add to Set.
3. Enter "Cardio" in the input box and click OK.
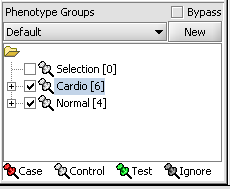
4. Next, similarly label the follow arrays as "Normal" ( repeat steps 2 & 3 ):
JB-n_0106.txt , JB-n_0821.txt, JB-n_0915.txt, JB-n_1303.txt
Activating sets
5. Select the checked boxes next to the panel name to indicate that these sets of arrays are "Active". Various analysis and visualization components can be set to only use/display activated arrays or markers.
Classifying a set
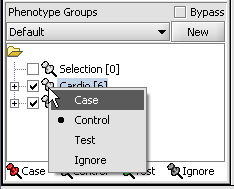
For statistical tests such as the t-test, Case and Control groups can be specified.
- Left-click on the thumb-tack icon in front of the phenotype name.
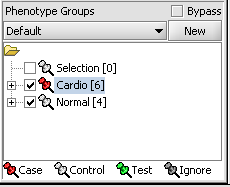
- Select Case to specify the disease arrays as the "Case". The remaining "Normal" arrays are by default labeled control.
A red thumbtack indicates the arrays have been specified as "Case".